Received 15 December 2003 | Accepted 11 June 2004 | Published 17 June 2004
Reprint
Received 15 December 2003 | Accepted 11 June 2004 | Published 17 June 2004 |
Download Reprint |
Differentially expressed genes in the lens of mimecan-null mice
Elena S. Tasheva,1 An Ke,1
Youping Deng,1 Chen Jun,1 Larry J. Takemoto,1 Anja
Koester,2 Gary W. Conrad1
1Kansas State University, Division of Biology, Manhattan, KS; 2Lilly Research Laboratories, Eli Lilly & Co., Indianapolis, IN
Correspondence to: Elena S. Tasheva, Division of Biology, Ackert Hall, Kansas State University, Manhattan, KS, 66506-4901; Phone: (785) 532-6553; FAX: (785) 532-6653; email: est@ksu.edu
Abstract
Purpose: Members of the small leucine-rich proteoglycans (SLRP) gene family are essential for normal collagen fibrillogenesis in various connective tissues and important regulators of cellular growth, differentiation, and tissue repair. Mimecan is a member of this gene family and is expressed in many connective tissues. We have previously reported that knockout of the mouse mimecan gene results in abnormal collagen fibrillogenesis, mainly in the cornea and skin. During the course of our studies on biological roles of mimecan in the eye, we found that this gene is expressed in the mouse lens. Here, we sought to identify gene expression changes in the lens that are associated with the absence of mimecan.
Methods: Reverse transcription-polymerase chain reaction amplification (RT-PCR), in situ hybridization (ISH), and immunohistochemistry (IHC) were used to determine mimecan expression in human and mouse eyes. Microarray hybridization was used to determine gene expression differences between lenses isolated from mimecan-null and wild type mice. Relative quantitative RT-PCR was used to verify the expression levels of a subset of the identified genes.
Results: By ISH and IHC, mimecan mRNA was detected in cornea and lens at embryonic day 16.5 (E16.5) and postnatal day 10 (P10) mouse eyes. By RT-PCR, mimecan mRNA was detected in human cornea, lens, iris, and retina. In mimecan-null mice lenses, microarray analysis of 5,002 mouse genes demonstrated a more than two fold increase in expression of 65 genes and a more than two fold decrease in expression of 76 genes. Among genes with increased expression were cell adhesion molecules, G-protein coupled receptors, intracellular signaling molecules, genes involved in protein biosynthesis and degradation, and genes involved in immune function. Decreased expression was found in extracellular matrix molecules, calcium binding and transporting proteins, and genes known for their roles in regulating cellular motility. Intriguingly, decreased gene expression was observed with two SLRP family members, biglycan and condroadherin, as well as with several stress-response proteins, including γA-crystallin, hemoglobin alpha 1, and metallothionein 1. Quantitative RT-PCR confirmed changes in expression of 12 genes selected from the arrays.
Conclusions: In this report we present the first demonstration that mimecan is constitutively expressed in the vertebrate lens. The results from gene expression profiling reveal the ability of mimecan to influence expression of biglycan and chondroadherin, thereby indicating possible novel regulatory interactions between these SLRP family members. As with mimecan, the expression of chondroadrein in vertebrate lens has not been reported previously. Our results provide insight into the function of mimecan in the lens and enable further characterization of molecular mechanisms by which this protein exerts its biological roles.
Introduction
Proteoglycans (PGs) of the extracellular matrix long have been recognized as organizers of collagenous networks and also as molecules that exhibit cell signaling properties, thereby influencing cellular growth, differentiation, and migration [1,2]. Among extracellular PGs, SLRPs form a rapidly growing subfamily currently including 13 members. Mimecan, a member of this gene subfamily, was isolated initially as a glycoprotein of 12 kDa from bovine bone and later as a 25 kDa keratan sulfate-containing PG from bovine cornea. The protein of 12 kDa was originally named osteoinductive factor and subsequently renamed osteoglycin [3-5]. The name mimecan was given to the full-length, 34 kDa, protein [6].
In vitro studies indicate that mimecan might play role(s) in many biological processes, including cellular growth, angiogenesis, and inflammation. This notion is supported by the following seven observations. (1) The genomic structure of mimecan is highly conserved between species; a single copy gene gives rise to multiple mRNA transcripts, three of which result from differential splicing within the first translated exon [7-9]. However, all mimecan mRNAs produce an identical protein that is conserved between mice, bovine, and man, suggesting its functional importance. (2) Transcriptional control elements are conserved between species. An E-box element is present in the proximal promoter region of human, bovine, rat, and mouse mimecan genes [10-12]. A p53 DNA-binding site also is conserved between these species [11]. Transcription factors that bind these elements, upstream stimulatory factor (USF) to the E-box and tumor suppressor protein p53 to its corresponding site, are important regulators of cellular growth and are activated in response to cellular stress. These observations suggest that mimecan may play roles in the same cellular processes in which p53 and USF are involved, i.e. adaptive responses, growth control, apoptosis, and ageing. (3) Expression of mimecan is tightly regulated in normal cells. Levels of mimecan mRNAs are high in corneal keratocytes maintained in low serum/serum-free media, but rapidly decrease if cells are grown in media containing serum [13]. (4) Growth factors and cytokines modulate mimecan mRNA expression in bovine and human cells [7,14-16]. (5) This gene is up-regulated after vascular injury and after low-level laser irradiation of osteoblasts, indicating that the corresponding protein may play a role in wound healing in vascular smooth muscle cells and in osteoblasts [14,17]. (6) Mimecan mRNA is up-regulated in activated endothelium and neo-intima in atherosclerotic lesions, observations suggesting roles for this protein in atherosclerosis [18]. (7) In most cancer cell lines and tumors mimecan is absent or is expressed at low levels [11].
Knockout of the mouse mimecan gene in vivo leads to increased collagen fibril diameters in cornea and skin, illustrating a role in collagen fibrillogenesis [19]. Notably, all single or double SLRP-null mice generated so far display abnormal collagen fibrillogenesis. As a result, these mice develop a variety of diseases such as osteoporosis, osteoarthritis, muscular dystrophy, Ehlers-Danlos syndrome, and corneal pathology [20-23]. However, besides the possibly redundant roles of the SLRPs in collagen fibrillogenesis, data also illustrate additional, unique roles of each one of these genes in other biological processes, such as cellular growth control and wound healing [1,2].
During the course of our studies on biological roles of mimecan in the eye, we found that that this gene is expressed not only in the cornea and sclera, but also in the iris, lens, and retina. Here we present the first demonstration that mimecan is constitutively expressed in mouse lens. To uncover potential biological roles of mimecan in the lens, we examined gene expression changes in the lens of mimecan-null mice using microarray technology. We show that 65 transcripts were increased and 76 transcripts were decreased by two fold or more in the absence of mimecan. Changed expression was found in genes encoding extracellular matrix components, G-protein-coupled signaling molecules, calcium-sensitive components of the cytoskeleton, and stress response proteins. Results presented here are an essential prerequisite for future functional studies aimed at an accurate understanding of molecular mechanisms by which mimecan exerts its biological roles.
Methods
RNA isolation and microarray analysis
Whole human eyes obtained from organ donors within 24 h post-mortem were provided by the Missouri Lions Eye Bank, Columbia, MO. Total RNA from cornea, iris, lens, and retina was isolated using Totally RNA Kit according to manufacturer's instructions (Ambion, Inc., Austin, TX).
Mice were housed in animal care facilities according to NIH guidelines (NIH publication number 86-23,1985) and IACUC approved protocols. All experiments were performed in compliance with ARVO statement for use of animals in ophthalmic and vision research.
Mouse lenses were obtained and immediately frozen in liquid nitrogen until isolation of RNA. For RNA isolation, probe synthesis and labeling, array hybridization and scanning the BD AtlasTM Custom Plastic Hybridization & Analysis Service (BD Biosciences Clontech, Palo Alto, CA) was used. One pool of total RNA samples was prepared from lenses of four 4 month old wild type mice. A separate pool was prepared from lenses of four age-matched mimecan-null mice. RNA quality and quantity were assessed by UV spectroscopy and capillary electrophoresis. Radioactive probes synthesized from these RNA pools were hybridized to three separate Clontech Atlas Plastic Mouse 5K Microarrays. The scanned images were analyzed using BD AtlasImageTM 2.7 software according to user guidelines. Briefly, background signal intensities were calculated using the default external background method (the median intensity of the "blank space" between the different panels of the array). For a small number of spots that were affected by signal bleed or where the signal was ambiguous, the background was estimated manually. The background signal was used to determine the signal intensities of individual spotted genes. For spots that were manually adjusted, the genes were excluded from the analysis, though the spots were used to normalize the signal intensity between the arrays. The Global Sum Normalization method was used to normalize the signal intensity between the experimental and control arrays. This method adds the values of signal over background for all genes on the arrays to calculate a normalization coefficient:
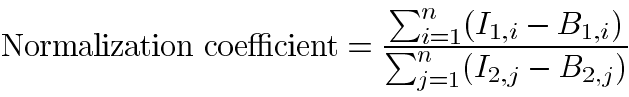
where Ix,y is the intensity of spot y on array x, Bx,y is the background intensity of spot y on array x, and n is the number of genes on the arrays. Adjusted intensities were computed as the intensity minus the background value of the gene, multiplied by the normalization coefficient.
Genes were considered to be differentially expressed if there was at least a two fold difference, either an increase or a decrease, in expression levels between wild type and knockout samples (normalized spot intensity knock-out sample:normalized spot intensity wild type sample >2.0). The raw data from these microarrays can be accessed in Appendix 1.
Statistical analysis
The data from three independent microarray hybridization experiments were analyzed using GeneSpring Version 6.1 software (Silicon Genetics, Redwood City, CA). The 141 differentially expressed genes were selected by two criteria; two fold change and one-way ANOVA. Differences were considered statistically significant when p values were <0.05.
Relative quantitative RT-PCR
Relative quantitative PCR followed by agarose gel electrophoresis was used to verify differentially expressed genes. Reverse transcription was carried out as previously described [8]. Quantum RNA18S Internal Standards (Ambion, Catalog numbers 1716 and 1717 amplifying 488 and 324 bp fragments, respectively) were used as endogenous standards in all PCR reactions. A 3:7 ratio of 18S primers to 18S competimers was added to 50 μl PCR amplification reactions containing 1X PCR buffer (Promega Corporation, Madison, WI), 5 units Taq polymerase (Promega), 0.2 mM dNTPs, and 100 ng of each gene specific primer pair. PCR reactions were programmed for 25 cycles (95 °C, 1 min; 60 °C, 1 min; 72 °C, 2 min). Resulting PCR products were analyzed by electrophoresis on 3.0% agarose gels, with DNA visualized by ethidium bromide staining. Sequences of all gene-specific primers used for verification of the microarray data are shown in Table 1. The PCR products that have been amplified using mouse lens RNA and that corresponded to mouse mimecan, chondroadherin, biglycan, and troponin I were cloned into pGEM-T and sequenced in both directions to confirm band specificity.
Immunohistochemisry
Custom IHC on slides containing normal adult mouse tissues was performed by SuperBioChips Laboratories, Seoul, Korea. Briefly, sections of mouse tissues were immunostained with the rabbit anti-osteoinductive factor polyclonal antibody raised against synthetic peptide corresponding to amino acids 206-217 of human mimecan (Chemicon International). Biotin-conjugated secondary antibody was reacted with avidin-biotin-peroxidase complex solution (Vector Laboratories Inc., Burlingame, CA), with immunoreactivity developed in 3,3'-diaminobenzidine (DAB).
In situ hybridization
For the ISH experiments, the custom service from Phylogeny Inc. was used. Briefly, mice tissues were fixed in 4% paraformaldehyde in phosphate-buffered saline overnight, dehydrated, and infiltrated with paraffin. Thin sections were mounted on gelatinized slides, deparaffinized in xylene, rehydrated, and post-fixed. The sections were digested with proteinase K, post-fixed, treated with triethanolamine/acetic anhydride, washed, and dehydrated. Sections were hybridized overnight at 53 °C in solutions that contained 50% deionized formamide and 50-75,000 cpm/μl 35S-labeled cRNA probe. The tissue was subjected to stringent washing and treated with 20 μg/ml RNAse A at 37 °C for 30 min. The slides were washed, dehydrated, dipped in Kodak NTB-2 nuclear track emulsion, and exposed for 1-2 weeks in light tight boxes with dessicant at 4 °C.
Bovine mimecan cDNA was cloned into pGEM-T vector and used as template for cRNA probe synthesis as previously described [8]. The cRNA probes were synthesized according to the manufacturer's conditions (Ambion) and labeled with 35S-UTP (>1,000 Ci/mmol, Amersham).
Photographic development was carried out in Kodak-D-19. Slides were counterstained with toluidine blue and analyzed using both light- and darkfield optics of a Zeiss Axiophot microscope. The slides were scanned into Photoshop 6.0 TIFF files.
Results
Mimecan is expressed in mouse lens
A search of the UniGene database at the NCBI showed that mimecan is expressed in almost all parts of human and mouse eyes, albeit at different levels. Thus, its sequence is present in several ESTs derived from cDNA libraries constructed from fetal eyes, lens, eye anterior segment, optic nerve, retina, foveal and macular retina, retinal pigment epithelium, and choroid (BM715748, BM701204, BM674101, BU741308, and BM724629), in a library from human retinal pigment epithelium/choroid cDNA (CA396423), in a library from human trabecular meshwork cDNA (CD676750, CD678059, and CD678058), ESTs from mice whole eye (CD808249, CD806521, and CB847562), and from mouse retina (GenBank Accession number BG296072). To confirm the expression patterns of mimecan in human eyes, we used RT-PCR. Primers were designed to flank an intron and to detect two major, differentially spliced mRNA species (Figure 1). In addition to the cornea, mimecan mRNA was detected in iris, lens, and retina. However, in contrast to cornea, skeletal muscle, brain, and placenta, in which two differentially spliced mRNA species were detected, only a single mRNA species was amplified in iris, lens, and retina.
To examine the expression of mimecan in mouse eye, we used in situ hybridization. Mimecan expression was detected in the lens and cornea of the E16.5 and P10 mouse eyes. Digital images of sections that show these results are presented in Figure 2.
Mimecan also was detected in mouse lens by IHC (Figure 3). Anti-mimecan antibody raised against amino acids 206-217 of human mimecan was used for these studies. As shown in Figure 3, this antibody could detect mimecan in mouse cartilage, thus demonstrating its ability to interact not only with human but also with mouse mimecan. By IHC the level of mimecan in mouse lens was found low compared to the high level of this protein in laryngeal cartilage (Figure 3). Taken together, these data demonstrate that mimecan is constitutively expressed in the vertebrate lens.
Differentially expressed genes identified by microarray analysis
Pooled target RNAs were used to perform hybridization on three separate microarrays. The Atlas Plastic Mouse 5K microarray is composed of 5002 full-length and EST (expressed sequence tag) cDNAs spotted in duplicate. A catalog of all genes spotted on this array, along with their GenBank accession numbers, is available at the BD Biosciences Clontech web site. The microarray also contains the cDNA Synthesis Control Spots, a phage lambda DNA sequence that has been spotted on the microarray to serve as positive control when used in conjuction with the cDNA Synthesis Control. The cDNA Synthesis Control consists of a synthetic RNA, corresponding to the sequence printed on the microarray. This control is spiked into the experimental RNA sample along with specific cDNA Synthesis Primers and is used to monitor the cDNA synthesis reaction. Successful cDNA synthesis results in adequate labeling and hybridization to the Atlas Plastic Microarray (Appendix 1).
The three hybridization experiments produced results consistent with one other (Appendix 1). The correlation coefficients between two samples within each group are above 0.89, whereas correlation coefficients between two samples from two different groups are less than 0.5 (Table 2). The hierarchy Clustering Method (GeneSpring, version 6.1) confirmed these correlation computations. The hierarchical clustering and scatter-plots of gene expressions are shown in Figure 4 and Figure 5. Of 5,002 genes represented on the Atlas Plastic Mouse 5K Microarray, 141 displayed greater than two fold expression changes. The 65 up-regulated and 74 down-regulated genes are listed in Table 3 and Table 4, respectively.
Validation of microarray data by relative quantitative PCR
To confirm the results of the microarray data, relative quantitative PCR was performed with the same RNA samples used for the microarray hybridizations. Twelve genes whose expression was altered in the absence of mimecan were selected for this confirmation analysis. Three of these genes (opsin, recoverin, and guanine nucleotide binding protein [G protein] gamma 3 subunit) exhibited increased expression in mimecan-null lenses based on the microarray data. Nine genes, including chondroadherin, biglycan, troponin C, troponin I, hemoglobin alpha, calcium transporting ATPase, keratin complex 1, apolipoprotein D, and γA-crystallin, exhibited decreased expression in mimecan-null lenses based on the microarray data. Two genes, γC-crystallin and γD-crystallin, that did not exhibit altered expression changes in the absence of mimecan, were also included as additional controls. Relative quantitative PCR yielded results consistent with microarray data for all twelve genes (Figure 6). Scanning densitometry of the gels containing ethidium bromide stained PCR products confirmed the trends in expression pattern of these 12 genes revealed by the microarray data (not shown).
Discussion
Members of the SLRP gene family are thought to be expressed mainly in cells of mesenchymal origin. However, their transient expression during the process of epithelial to mesenchymal cell transition in response to injury, such as cataract surgery, also has been documented [24]. Our data demonstrate that mimecan, a SLRP family member, is constitutively expressed in the lens. Mimecan mRNA also was detected in cultured bovine lens epithelial cells by PCR (not shown). By IHC we found significantly reduced mimecan, in mouse lens (compare intensity of the brown color between lens, sclera, and two cartilage samples in Figure 3), whereas ISH shows high levels of mimecan mRNA in the lens (Figure 2). At this time we do not know the reason for this discrepancy between RNA and protein levels. However, several factors could contribute to this finding. First, it is possible that mimecan mRNA levels are abundant in the lens because this RNA may have function(s) other than serving as a template for protein synthesis. In support of this hypothesis is the observation that multiple mimecan mRNA splice variants are synthesized in many cell types, yet the protein remains unchanged. Secondly, mimecan protein turnover may be high in the lens. Thirdly, a combination of differential RNA abundance and differential translation in epithelial and fiber cells may be the explanation for these data.
Our finding that mimecan is constitutively expressed in the lens suggests that this protein (or its mRNA) may have a specific role in maintaining lens morphology and physiology. We therefore used microarray analysis to investigate how the absence of mimecan would affect the gene expression profile of mouse lens. Based on our previous demonstration that mimecan is expressed in cornea and also in sclera of several mammalian species [6], as well as on data obtained by searching UniGene databases showing mimecan expression in almost all parts of the eye, initially we performed microarray analysis using RNA isolated from whole mouse eyes. Due to the limitation of this approach (results obtained represent the average expression of a gene for all cell types in the eye) we subsequently chose lens tissue for this analysis. Lens tissue contains only two cell types, epithelial and fiber cells, which makes the results easier to comprehend. In addition, such results would provide important new information for deciphering the function of mimecan in the lens. The results demonstrate that targeted disruption of the mimecan gene in the mouse leads to changes in expression of several important components of the lens. They also suggest at least two potential mechanisms by which mimecan may exert its biological function in the lens; (i) by binding cell surface receptor(s), similar to other extracellular matrix proteins [25], thereby triggering signaling pathway(s) that cause global changes in lens gene expression, or (ii) by remaining inside lens cells (similar to biglycan in neuronal cells [26,27]) and interacting directly with intracellular protein(s).
We also confirmed the expression levels of a subset of the identified genes by relative quantitative PCR. Of note, the results from this study, showing that the expression of a number of genes is affected by the absence of mimecan, are interesting in view of the fact that initial examination of mimecan-null mice did not reveal major developmental changes of the lens. However, lack of phenotypic changes has been reported for many other genes known to be expressed in the lens [28-30]. A lack of major phenotypic changes may indicate either functional redundancy with other proteins or the need to knock-out the gene of interest in a different mouse background. We also should note that lens morphology was not monitored carefully during our initial characterization of mimecan-null mice. Focus was given mainly to the cornea, because at that time we did not have data demonstrating mimecan expression in the lens. Ultrastructural analysis of lenses at various ages of mimecan-null mice also has not been performed. Future studies will be aimed at addressing roles of mimecan in lens morphology.
The functions of known genes found differentially expressed in the absence of mimecan corresponded well with our data that demonstrate a role of mimecan in collagen fibrillogenesis, as well as with our data on transcriptional regulation of mimecan that suggest a role for this protein in cellular growth, motility, and stress response [10-12,16,20].
The genes found to be differentially expressed in the lens of mimecan-null mice can be grouped into several functional categories, including extracellular matrix and adhesion molecules, G-protein coupled receptors, calcium-sensitive components of the cytoskeleton, stress proteins, metabolic enzymes, and transcription factors (Table 3 and Table 4).
The observation that among extracellular matrix genes found differentially expressed in the mimecan-null lenses were chondroadherin and biglycan is very interesting because it suggests possible novel regulatory interactions between these SLRP family members. Furthermore, finding that chondroadherin also is expressed in mouse lens is novel. Chondroadherin belongs to Class IV of the SLRP gene family and is known as a cartilage protein [31,32]. It binds to chondrocytes, fibroblasts, and osteoblasts via integrin α(2)β(1) receptors and thus promotes cell attachment [25]. Chondroadherin also binds to collagens type II and type VI [33,34]. Biglycan is a well characterized member of the SLRP gene family, shown to regulate collagen fibrillogenesis [20,21]. Biglycan also binds TGF-β and has been found in the nuclei of neuronal cells, where it may be involved in the regulation of cellular proliferation [26,27]. Changed expression of matrix metalloproteinase-2 and tissue inhibitor of metalloproteinases in the absence of mimecan suggests a role of this protein in tissue remodeling.
Among genes that belong to the G-protein coupled receptor protein signaling pathway are opsin1 short-wave sensitive (16.4 fold increase), opsin 1 medium-wave sensitive (22.6 fold increase), several guanine nucleotide binding and transducing proteins, and phosphodiesterase 6G and 6D (Table 3). G-protein coupled receptors activate phospholipase Cβ, thereby generating the calcium-mobilizing second messenger molecules leading to calcium release into the cytoplasm [35]. Because of the involvement of G-protein coupled receptor signaling molecules in numerous physiological and pathological conditions, they are being increasingly targeted to treat a range of ocular problems (reviewed in [35]). How mimecan affects the expression of these genes remains to be determined.
Several genes encoding important components of the cytoskeleton that play roles in cell motility and muscle contraction and that are calcium-sensitive were suppressed in the absence of mimecan, including S100 calcium binding proteins A10 and A13, troponin C, I and T3, tropomyosin, creatine kinase, enolase 3, calsequestrin 1, calcium transporting ATPase, annexins A1, A2, and A4, calpain, prothymosin beta 4, and recoverin (Table 4). Troponin is the thin filament regulatory complex that confers calcium sensitivity to striated muscle actomyosin ATPase activity and is composed of three polypeptides, troponin I, troponin T, and troponin C. Troponin I is the subunit that in the presence of tropomyosin inhibits myosin Mg2+-ATPase activity. Troponin C is the Ca2+-sensitive component of the complex [36]. In addition to its role in muscular contractions, troponin I is a potent inhibitor of angiogenesis and tumor metastasis [37]. Troponin I was found suppressed by 11.2 fold in the absence of mimecan. Given the observation that troponin I is also expressed in cartilage where it suppresses blood vessel formation, its suppression in the mimecan-null lens supports the hypothesis that mimecan may play role in lens transparency. Creatine kinase (CK) is an abundant enzyme, important for maintenance of high-energy phosphate homeostasis in many tissues. Double-knockout CK mice, missing both the muscle and sarcomeric mitochondrial isoforms of CK, display large changes in skeletal muscle function [38]. Beta enolase is the beta subunit of the glycolytic enzyme, enolase. Enzyme enolase catalyzes the interconversion of 2-phosphoglycerate and phosphoenolpyruvate. Muscle beta-enolase deficiency has been considered in differential diagnosis of metabolic myopathies due to inherited defects of distal glycolysis [39]. Calcium-binding protein A11, calgizzrain, is a member of the S100 family of proteins. They are localized in the cytoplasm and/or nucleus of a wide range of cell types and are involved in the regulation of cell differentiation and motility, tubulin polymerization, and tumor invasion [40,41]. Sarcoplasmic reticulum Ca2+-ATPase1 (ATP2A1) is a 110-kDa membrane protein that catalyzes the ATP-dependent transport of Ca2+ from the cytosol to the lumen of the sarcoplasmic reticulum [42]. Defects in the ATP2A1 gene are the cause of Brody disease, a rare inherited disorder of skeletal muscle, resulting in exercise-induced impairment of skeletal muscle relaxation, stiffness, and cramps [43]. Targeted disruption of the ATP2A1 gene impairs diaphragm function and is lethal in mice [44]. Suppression of such a large group of calcium-sensitive genes is an important observation in view of the fact that increased lens cell calcium is known to trigger the activation of calpain, a protease that may be responsible for the modification of cytoskeletal proteins and crystallins in lens cataract models (reviewed in [35]). This observation strongly suggest a potential role of mimecan in the maintenance of normal lens transparency.
γA-Crystallin and hemoglobin alpha were suppressed by 2.5 and 12.5 fold, respectively. Crystallins are known to account for 80-90% of water-soluble proteins of the transparent lens, where they contribute to the optical properties of the lens [45,46]. Recently, several isotypes of hemoglobin have been shown expressed in the lens [47]. Because the role of hemoglobin in lens is largly unknown, availability of mimecan-null mice will provide a useful model for studying the roles of this protein as well.
The absence of mimecan also led to altered expression of several genes involved in cellular immune responses, such as interleukin 15, lymphocyte antigen 6a, and STAT 6. Most of these genes are ubiquitously expressed and also have functions other than immune response. The expression of genes associated with protein synthesis, defense against oxidative stress, and chaperone activity also was altered in mimecan-null lenses. These include cytochrome c oxdase, glutatione S-tranferase, glutatione peroxidase, aldehyde dehydrogenase, metallothionein 1, and high mobility group nuclosomal protein 1 (Table 3 and Table 4). Altered expression of these genes have been associated with different types of cataract [48].
Taken together, results from this study indicate involvement of mimecan in several physiological and pathological processes in vertebrate lens and provide new information that may lead to development of new therapies for ocular diseases.
Acknowledgements
This work was supported by NIH Grant EY13395 to GWC and EST. We thank Dr. Ron Walkenbach, Tina Livesay, and Amy Giangiacomo, Missouri Lions Eye Bank for the human eyes.
References
1. Iozzo RV. The biology of the small leucine-rich proteoglycans.
Functional network of interactive proteins. J Biol Chem 1999;
274:18843-6. ![]()
2. Kresse H, Schonherr E. Proteoglycans of the extracellular matrix
and growth control. J Cell Physiol 2001; 189:266-74. ![]()
3. Bentz H, Nathan RM, Rosen DM, Armstrong RM, Thompson AY, Segarini
PR, Mathews MC, Dasch JR, Piez KA, Seyedin SM. Purification and
characterization of a unique osteoinductive factor from bovine bone. J
Biol Chem 1989; 264:20805-10. ![]()
4. Dasch JR, Pace DR, Avis PD, Bentz H, Chu S. Characterization of
monoclonal antibodies recognizing bovine bone osteoglycin. Connect
Tissue Res 1993; 30:11-21. ![]()
5. Madisen L, Neubauer M, Plowman G, Rosen D, Segarini P, Dasch J,
Thompson A, Ziman J, Bentz H, Purchio AF. Molecular cloning of a novel
bone-forming compound: osteoinductive factor. DNA Cell Biol 1990;
9:303-9. ![]()
6. Funderburgh JL, Corpuz LM, Roth MR, Funderburgh ML, Tasheva ES,
Conrad GW. Mimecan, the 25-kDa corneal keratan sulfate proteoglycan, is
a product of the gene producing osteoglycin. J Biol Chem 1997;
272:28089-95. ![]()
7. Tasheva ES, Funderburgh ML, McReynolds J, Funderburgh JL, Conrad
GW. The bovine mimecan gene. Molecular cloning and characterization of
two major RNA transcripts generated by alternative use of two splice
acceptor sites in the third exon. J Biol Chem 1999; 274:18693-701.
![]()
8. Tasheva ES, Corpuz LM, Funderburgh JL, Conrad GW. Differential
splicing and alternative polyadenylation generate multiple mimecan mRNA
transcripts. J Biol Chem 1997; 272:32551-6. ![]()
9. Ujita M, Shinomura T, Kimata K. Molecular cloning of the mouse
osteoglycin-encoding gene. Gene 1995; 158:237-40. ![]()
10. Tasheva ES. Analysis of the promoter region of human mimecan
gene. Biochim Biophys Acta 2002; 1575:123-9. ![]()
11. Tasheva ES, Maki CG, Conrad AH, Conrad GW. Transcriptional
activation of bovine mimecan by p53 through an intronic DNA-binding
site. Biochim Biophys Acta 2001; 1517:333-8. ![]()
12. Tasheva ES, Conrad GW. The UV responsive elements in the human
mimecan promoter: a functional characterization. Mol Vis 2003; 9:1-9
<http://www.molvis.org/molvis/v9/a1/>. ![]()
13. Beales MP, Funderburgh JL, Jester JV, Hassell JR. Proteoglycan
synthesis by bovine keratocytes and corneal fibroblasts: maintenance of
the keratocyte phenotype in culture. Invest Ophthalmol Vis Sci 1999;
40:1658-63. ![]()
14. Shanahan CM, Cary NR, Osbourn JK, Weissberg PL. Identification of
osteoglycin as a component of the vascular matrix. Differential
expression by vascular smooth muscle cells during neointima formation
and in atherosclerotic plaques. Arterioscler Thromb Vasc Biol 1997;
17:2437-47. ![]()
15. Long CJ, Roth MR, Tasheva ES, Funderburgh M, Smit R, Conrad GW,
Funderburgh JL. Fibroblast growth factor-2 promotes keratan sulfate
proteoglycan expression by keratocytes in vitro. J Biol Chem 2000;
275:13918-23. ![]()
16. Tasheva ES, Conrad GW. Interferon-gamma regulation of the human
mimecan promoter. Mol Vis 2003; 9:277-87 <http://www.molvis.org/molvis/v9/a39/>.
![]()
17. Hamajima S, Hiratsuka K, Kiyama-Kishikawa M, Tagawa T, Kawahara
M, Ohta M, Sasahara H, Abiko Y. Effect of low-level laser irradiation on
osteoglycin gene expression in osteoblasts. Lasers Med Sci 2003;
18:78-82. ![]()
18. Fernandez B, Kampmann A, Pipp F, Zimmermann R, Schaper W.
Osteoglycin expression and localization in rabbit tissues and
atherosclerotic plaques. Mol Cell Biochem 2003; 246:3-11.
![]()
19. Tasheva ES, Koester A, Paulsen AQ, Garrett AS, Boyle DL, Davidson
HJ, Song M, Fox N, Conrad GW. Mimecan/osteoglycin-deficient mice have
collagen fibril abnormalities. Mol Vis 2002; 8:407-15 <http://www.molvis.org/molvis/v8/a48/>.
![]()
20. Ameye L, Young MF. Mice deficient in small leucine-rich
proteoglycans: novel in vivo models for osteoporosis, osteoarthritis,
Ehlers-Danlos syndrome, muscular dystrophy, and corneal diseases.
Glycobiology 2002; 12:107R-16R. ![]()
21. Ameye L, Aria D, Jepsen K, Oldberg A, Xu T, Young MF. Abnormal
collagen fibrils in tendons of biglycan/fibromodulin-deficient mice lead
to gait impairment, ectopic ossification, and osteoarthritis. FASEB J
2002; 16:673-80. ![]()
22. Jepsen KJ, Wu F, Peragallo JH, Paul J, Roberts L, Ezura Y,
Oldberg A, Birk DE, Chakravarti S. A syndrome of joint laxity and
impaired tendon integrity in lumican- and fibromodulin-deficient mice. J
Biol Chem 2002; 277:35532-40. ![]()
23. Corsi A, Xu T, Chen XD, Boyde A, Liang J, Mankani M, Sommer B,
Iozzo RV, Eichstetter I, Robey PG, Bianco P, Young MF. Phenotypic
effects of biglycan deficiency are linked to collagen fibril
abnormalities, are synergized by decorin deficiency, and mimic
Ehlers-Danlos-like changes in bone and other connective tissues. J Bone
Miner Res 2002; 17:1180-9. ![]()
24. Saika S, Miyamoto T, Tanaka S, Tanaka T, Ishida I, Ohnishi Y,
Ooshima A, Ishiwata T, Asano G, Chikama T, Shiraishi A, Liu CY, Kao CW,
Kao WW. Response of lens epithelial cells to injury: role of lumican in
epithelial-mesenchymal transition. Invest Ophthalmol Vis Sci 2003;
44:2094-102. ![]()
25. Camper L, Heinegard D, Lundgren-Akerlund E. Integrin alpha2beta1
is a receptor for the cartilage matrix protein chondroadherin. J Cell
Biol 1997; 138:1159-67. ![]()
26. Hildebrand A, Romaris M, Rasmussen LM, Heinegard D, Twardzik DR,
Border WA, Ruoslahti E. Interaction of the small interstitial
proteoglycans biglycan, decorin and fibromodulin with transforming
growth factor beta. Biochem J 1994; 302 (Pt 2):527-34. ![]()
27. Liang Y, Haring M, Roughley PJ, Margolis RK, Margolis RU.
Glypican and biglycan in the nuclei of neurons and glioma cells:
presence of functional nuclear localization signals and dynamic changes
in glypican during the cell cycle. J Cell Biol 1997; 139:851-64.
![]()
28. Dong L, Chen Y, Lewis M, Hsieh JC, Reing J, Chaillet JR, Howell
CY, Melhem M, Inoue S, Kuszak JR, DeGeest K, Chung AE. Neurologic
defects and selective disruption of basement membranes in mice lacking
entactin-1/nidogen-1. Lab Invest 2002; 82:1617-30. ![]()
29. Ebong S, Chepelinsky AB, Robinson ML, Zhao H, Yu CR, Egwuagu CE.
Characterization of the roles of STAT1 and STAT3 signal transduction
pathways in mammalian lens development. Mol Vis 2004; 10:122-31
<http://www.molvis.org/molvis/v10/a16/>. ![]()
30. Graw J, Loster J. Developmental genetics in ophthalmology.
Ophthalmic Genet 2003; 24:1-33. ![]()
31. Neame PJ, Sommarin Y, Boynton RE, Heinegard D. The structure of a
38-kDa leucine-rich protein (chondroadherin) isolated from bovine
cartilage. J Biol Chem 1994; 269:21547-54. ![]()
32. Shen Z, Gantcheva S, Mansson B, Heinegard D, Sommarin Y.
Chondroadherin expression changes in skeletal development. Biochem J
1998; 330 (Pt 1):549-57. ![]()
33. Mansson B, Wenglen C, Morgelin M, Saxne T, Heinegard D.
Association of chondroadherin with collagen type II. J Biol Chem 2001;
276:32883-8. ![]()
34. Wiberg C, Heinegard D, Wenglen C, Timpl R, Morgelin M. Biglycan
organizes collagen VI into hexagonal-like networks resembling tissue
structures. J Biol Chem 2002; 277:49120-6. ![]()
35. Duncan G, Collison DJ. Calcium signalling in ocular tissues:
functional activity of G-protein and tyrosine-kinase coupled receptors.
Exp Eye Res 2002; 75:377-89. ![]()
36. Gomes AV, Potter JD, Szczesna-Cordary D. The role of troponins in
muscle contraction. IUBMB Life 2002; 54:323-33. ![]()
37. Moses MA, Wiederschain D, Wu I, Fernandez CA, Ghazizadeh V, Lane
WS, Flynn E, Sytkowski A, Tao T, Langer R. Troponin I is present in
human cartilage and inhibits angiogenesis. Proc Natl Acad Sci U S A
1999; 96:2645-50. ![]()
38. Kernec F, Unlu M, Labeikovsky W, Minden JS, Koretsky AP. Changes
in the mitochondrial proteome from mouse hearts deficient in creatine
kinase. Physiol Genomics 2001; 6:117-28. ![]()
39. Comi GP, Fortunato F, Lucchiari S, Bordoni A, Prelle A, Jann S,
Keller A, Ciscato P, Galbiati S, Chiveri L, Torrente Y, Scarlato G,
Bresolin N. Beta-enolase deficiency, a new metabolic myopathy of distal
glycolysis. Ann Neurol 2001; 50:202-7. ![]()
40. Sakaguchi M, Miyazaki M, Inoue Y, Tsuji T, Kouchi H, Tanaka T,
Yamada H, Namba M. Relationship between contact inhibition and
intranuclear S100C of normal human fibroblasts. J Cell Biol 2000;
149:1193-206. ![]()
41. Tanaka M, Adzuma K, Iwami M, Yoshimoto K, Monden Y, Itakura M.
Human calgizzarin; one colorectal cancer-related gene selected by a
large scale random cDNA sequencing and northern blot analysis. Cancer
Lett 1995; 89:195-200. ![]()
42. MacLennan DH, Rice WJ, Green NM. The mechanism of Ca2+ transport
by sarco(endo)plasmic reticulum Ca2+-ATPases. J Biol Chem 1997;
272:28815-8. ![]()
43. Brody IA. Muscle contracture induced by exercise. A syndrome
attributable to decreased relaxing factor. N Engl J Med 1969;
281:187-92. ![]()
44. Pan Y, Zvaritch E, Tupling AR, Rice WJ, de Leon S, Rudnicki M,
McKerlie C, Banwell BL, MacLennan DH. Targeted disruption of the ATP2A1
gene encoding the sarco(endo)plasmic reticulum Ca2+ ATPase isoform 1
(SERCA1) impairs diaphragm function and is lethal in neonatal mice. J
Biol Chem 2003; 278:13367-75. ![]()
45. Piatigorsky J. Multifunctional lens crystallins and corneal
enzymes. More than meets the eye. Ann N Y Acad Sci 1998; 842:7-15.
![]()
46. Bhat SP. Crystallins, genes and cataract. Prog Drug Res 2003;
60:205-62. ![]()
47. Wride MA, Mansergh FC, Adams S, Everitt R, Minnema SE, Rancourt
DE, Evans MJ. Expression profiling and gene discovery in the mouse lens.
Mol Vis 2003; 9:360-96 <http://www.molvis.org/molvis/v9/a50/>. ![]()
48. Hawse JR, Hejtmancik JF, Huang Q, Sheets NL, Hosack DA, Lempicki
RA, Horwitz J, Kantorow M. Identification and functional clustering of
global gene expression differences between human age-related cataract
and clear lenses. Mol Vis 2003; 9:515-37 <http://www.molvis.org/molvis/v9/a65/>.
![]()