Received 23 December 2003 | Accepted 22 October 2004 | Published 28 December 2004
Reprint
Received 23 December 2003 | Accepted 22 October 2004 | Published 28 December 2004 |
Download Reprint |
Temporal changes of novel transcripts in the chicken retina following imposed defocus
Sibylle Ohngemach, Christine Buck,
Perikles Simon,
Frank Schaeffel,
Marita Feldkaemper
Section of Neurobiology, University Eye Hospital, University of Tuebingen, Tuebingen, Germany
Correspondence to: Marita Feldkaemper, Ph.D., Section of Neurobiology of the Eye, University Eye Hospital, University of Tuebingen, Calwerstrasse, 7/1 72076, Tuebingen, Germany; Phone: (+49) 7071/29-87424; FAX: (+49) 7071/29-5196; email: marita.feldkaemper@uni-tuebingen.de
Abstract
Purpose: Changes in retinal gene expression are one of the first steps in the signaling pathway underlying the visual control of eye growth. We tried to identify novel, yet unknown, genes, that alter their expression pattern following imposed defocus, wearing of diffusers, or during recovery from myopia. Sequences found earlier by differential display studies were applied to 5'-RACE and identified as 15 kDa selenoprotein P and prolidase. Moreover, we obtained more sequence information for a yet unidentified gene. We have studied the time course of expressions of these genes following lens or diffuser treatment.
Methods: Ten to 14 day old white leghorn chickens (4-7) were treated with a monocular +7 D or -7 D lenses for 2, 4, 6, or 24 h, or treated with monocular or binocular diffusers for 2, 4, or 6 h. Chickens of another group were allowed to recover from 4 days of diffuser wear for 4 h. Untreated chicks served as a control for contralateral eye effects. Following the extraction of retinal RNA, the relative expression of the three genes was determined by semi-quantitative real time PCR.
Results: We found a significant up regulation of selenoprotein P expression after 24 h of treatment with positive (+380%) or negative lenses (+387%) which was even more prominent in the contralateral untreated eyes (positive: +542%; negative: +786%). A rapid change in selenoprotein mRNA levels was induced by binocular diffuser wear for 2 h (+425%), whereas defocus blur in one eye led to an increase only after 6 h (+261%). There was a significant upregulation of prolidase mRNA after 24 h of treatment with positive (+75%) but not with negative lenses. Moreover, blur induced by diffusers resulted in a highly significant rise of prolidase mRNA levels after 4 h, both with monocular (142%) and binocular (106%) treatment. This is similar to what was found in the previous differential display (DD) screening of monocularly treated eyes. In contrast to the findings of the DD screening, the mRNA expression of the unknown gene remained unchanged both after hyperopic and myopic defocus. Again, blur induced by diffusers evoked the most prominent change after 6 h of binocular treatment. There were no significant alterations in the mRNA levels of the three investigated genes after 4 h of recovery from myopia that was induced by a 4 day period of diffuser treatment.
Conclusions: The mRNA expression of selenoprotein P, prolidase, and of the not yet identified gene (sequence 3) is clearly altered by retinal image degradation imposed by diffuser wearing and, in part, by defocus imposed by spectacle lenses. However, none of the candidates are regulated by the sign of imposed defocus, suggesting a role in retinal contrast processing.
Introduction
Refraction and eye growth are regulated by visual processing in the retina. Here, both the amount and the sign of imposed defocus are recognized and determine the sign of growth changes of the underlying sclera [1]. So far, little is known about the messengers that are released from the retina to induce these changes. Recently, it was shown that the immediate-early gene product ZENK, which is expressed in glucagon amacrine cells, is increased in these cells if chicks were treated with positive lenses and decreased if negative lenses or diffusers were worn [2,3]. There is also evidence for a potential role of glucagon itself as a stop signal in eye growth [4]. Gene expression in the retina, choroid, and sclera was investigated in recent studies. In tree shrews, the mRNA expression of scleral enzymes, such as matrix metalloproteinases (MMP) and tissue inhibitors of MMP (TIMP), were quantified [5]. Gene expression changes in the sclera were seen after 11 days of form deprivation and after 4 days of recovery from deprivation myopia. However, these candidates must be considered as effectors acting in the target tissue for eye growth, the sclera, rather than initial messengers in the signaling cascade. Ishibashi et al. [6] found an upregulation of crystallin mRNAs in a retina-RPE-choroid complex of chickens wearing diffusers. However, the role of crystallins in eye growth regulation is still unclear. In a differential display (DD) RT-PCR screening, Feldkaemper et al. [7] detected 17 retinal and choroidal gene sequences that displayed altered expression with visual stimulation that is known to induce refractive errors. Only one sequence could be identified by database search as the chicken cytochrome-c oxidase subunit I. Its mRNA expression was shown to be significantly upregulated after 6 h or one day of treatment with positive lenses, while there was only a trend of downregulation induced by negative lenses or diffusers. Given that cytochrome-c oxidase is an important energy delivering enzyme of the oxidative phosphorylation pathway, the observed changes may reflect a change in metabolic requirements of the growth processes per se.
Methods
5'-RACE
Fragments of gene sequences were taken from a previous DD study in chickens by Feldkaemper et al. [7]. A probable reason why it was not possible to identify the genes in database searches was that the DD method provides sequence information at the 3' end of the genes which is generally more variable among species due to its non-coding nature. A further complicating factor is that the chicken genome is only partly known. In order to extend the sequence information to the 5' end, we applied the method of rapid amplification of cDNA ends (SMARTTM 5'-RACE, Clontech, Palo Alto). The sensitivity and specificity of the touchdown method protocol allow for the amplification of rare transcripts. RNA from differently treated chickens was pooled in order to obtain all possible transcripts. Following the manufacturers' instructions, we used 1 μg of total RNA in a volume of 10 μl for first strand cDNA synthesis. Six gene specific primers (Thermo Hybaid, Ulm, Germany) were designed using the software PrimerDesign and checked with Primer Premier 5 (Premier Biosoft International, Palo Alto, CA). We used the Advantage 2 PCR Enzyme System (Clontech), which permits amplifications of long templates. The raced PCR products were assessed by agarose gel electrophoresis. With three out of six primer sets we did not succeed in obtaining a PCR product. The PCR reaction with primer sets 1 to 3 provided bands that were extracted from the gels (NucleoTrap Nucleic Acid Purification Kit, Clontech), and cloned (TOPO TA Cloning Kit, Invitrogen). The inserts were sized by colony PCR. Amplicons that showed the correct length acquired by the gel electrophoresis prior to cloning were automatically sequenced. One of the PCR products that was amplified with the primer pair 2 was directly sequenced without a cloning step. Sequences were analyzed using the BLASTn and BLASTp tools and the Translate and Six Frame Translation of Sequence tools. The results are shown in Table 1.
Treatment of animals for real time PCR
Male white leghorn chickens were raised in groups on a 12 h light/12 h dark cycle. They were treated according to the ARVO resolution for care and use of laboratory animals in ophthalmic and vision research. The treatment protocols were as follows: 10 to 14 day old chickens were unilaterally treated with a +7 D lens or a -7 D lens for different periods of time (2 h, 4 h, 6 h, and 24 h). Another group of the same age was treated with monocular or binocular diffusers, for different periods of time (2 h, 4 h, and 6 h). A third group was subjected to monocular occlusion from day 9-12, followed by a 4 h period of recovery from occlusion on day 13. Four to seven animals were used for each treatment. All animals were sacrificed at the same time around noon. The lenses were attached to velcro rings that were glued to the feathers around the eyes under a light ether anesthesia. The diffusers were hand made translucent plastic shells and were directly glued to the feathers around the eyes. One group of six untreated chickens served as control group.
Retina preparation
Animals were killed by an overdose of diethyl-ether and immediately enucleated. Eyes were cut with a razor blade perpendicular to the anterior-posterior axis about 1 mm posterior to the ora serata, at about 50% of the axial length. The anterior segment of the eye and the lens were discarded. The retina was isolated from the eye cup in ice chilled Ringer solution under visual control through a dissecting microscope. Pieces attached to retinal pigment epithelium (RPE) were discarded. The tissue was snap frozen in liquid nitrogen and stored at -70 °C until RNA extraction.
RNA isolation and cDNA synthesis
Total RNA was extracted from the homogenized retinas according to the manufacturers' instructions (RNeasy Mini Kit; Qiagen, Hilden, Germany) and digested with RNase free DNase-I (Roche, Mannheim, Germany). The quality and quantity of the RNA was assessed by obtaining the ratio of absorbance values at 260 and 280 nm, and by visualizing the RNA on agarose gels. Each RNA sample (1 μg) was reverse transcribed by SuperScriptTMII RNase H-Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) with 0.5 μg oligo (dT)15 Primer (Roche).
Semi-quantitative real time PCR
The gene expression levels of the target genes were normalized to the reference gene 18S rRNA (AF173612). We used two different forward primers for 18S rRNA-amplification. For normalization of the selenoprotein expression, a longer part of the 18S rRNA was chosen as reference gene (referred to as 18S rRNA (1) in Table 2) whereas for normalization of prolidase and the sequence 3, a shorter part of the 18S rRNA was amplified (referred to as 18S rRNA (2) in Table 2) due to a better fit of the amplification efficiencies. Based on sequence information that was obtained after applying the 5'-RACE method, gene specific primers were selected. Their characteristics are also shown in Table 2. The product size was verified by agarose gel electrophoresis and automated sequencing. One of the identified sequences, guanine nucleotide binding protein β4, could not be quantified by real time PCR even though we used two different sets of primers. There might have been steric reasons for this failure. Also, several optimization efforts concerning annealing temperature, primer concentration, or template amount failed to improve the result, possibly due to an intron sequence that could be present only in chicks but not in humans or mice.
Real time PCR was carried out in the iCycler iQTM Multi-Color Real Time PCR Detection System (Bio-Rad, Hercules, CA) with the QuantiTect SYBR Green PCR Kit (Qiagen). Primer concentration was optimized to a final concentration of 0.6 μM and combined with 1 ng of retinal RNA per well. We set up 3 reactions per sample RNA (triplets) with a final volume of 15 μl per single reaction. Real time PCR was performed as a three step cycling under the following conditions: initial denaturation at 95 °C for 15 min; denaturation, annealing, and extension at 94 °C for 15 s, 58 °C for 30 s, and 72 °C for 1 min (40 cycles). Fluorescence was measured at 58 °C at the end of every cycle.
Data analysis and statistics
The evaluation of the real time PCR results is based on "threshold cycles" (CTs) for each PCR product. The CT value is defined as the PCR cycle at which the fluorescence intensity of a sample crosses a threshold line that is defined by the experimenter in the exponential phase of the amplification. The earlier the threshold is crossed, the smaller is the CT value and the higher is the amount of initial RNA. In order to obtain amplification efficiencies of the five primer sets, we generated standard curves by a twofold dilution series with template amounts ranging between 8 ng and 0.125 ng RNA per well (Figure 1). The efficiency (E) of the PCR reaction was calculated for each primer pair according to the equation E=10(-1/slope). A duplication of the logarithm of the template amount between two dilution steps results in an efficiency of 100%, resulting in E=2. With zero duplication (0%), the E would be 1. The slope (S) was determined by plotting the mean CT of each dilution as a function of the logarithm of the template amount. The slopes, efficiencies, and correlations of the standard curves are shown in Table 3.
To consider the variability of the PCR amplification of each primer pair we calculated the coefficient of variation (CV) by dividing the standard deviation (SD) of each triplet by the mean CT. In Table 3, the mean CV±SD of all triplets of reference and target genes are presented.
For statistical analysis and graphical presentation of the results we determined the mean normalized expression (MNE) [8] for each treatment group according to the following equation:
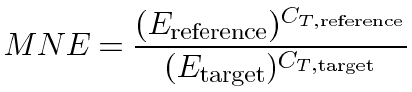
where E is the efficiency and CT is the mean threshold cycles of the PCR reaction. The "reference" subscript refers to the reference gene 18S rRNA (sequences 1 and 2) and the "target" subscript refers to the target genes selenoprotein, prolidase, and sequence 3.
For the statistical analysis of the time course (2, 4, 6, and 24 h) of gene expression for the different treatments (+7 D lens, -7 D lens, monocular diffuser, binocular diffuser), we applied separate one way ANOVAs with Tukey-Kramer and Dunnetts' tests for post hoc analysis. In the event that an ANOVA was significant, Tukey-Kramer test was applied to show differences in gene expression after various treatment periods within each treatment group and Dunnetts' test to compare each treatment group with the untreated control group. Moreover, all tests were done separately for untreated contralateral control eyes.
For an a priori comparison of the MNE of the treated and the untreated contralateral eyes, we used the paired Students' t-test with Bonferroni correction. Gene expression in eyes treated with monocular and binocular diffusers were compared separately for every treatment time using the unpaired, Bonferroni corrected Students' t-test. Since there was no correlation between both eyes in six untreated control animals, the MNE of the twelve single eyes were pooled.
Results
Changes in mRNA expression in the retina induced by lens or diffuser wear
There was an upregulation of selenoprotein mRNA of about 380% after 24 h of treatment with positive lenses (Figure 2) which was statistically significant (Table 4; Dunnetts' test: TE +7 D/CG: p<0.001). At this time point, the upregulation of selenoprotein mRNA content was even more pronounced in the untreated contralateral eye (+542% compared to the untreated control group: ANOVA: UE/CG: p<0.0008). All significant results are shown in Table 4. The differences between the treated eyes (TE) and the untreated contralateral eyes (UE) did not achieve significance at any of the time points.
The result was similar after treatment with negative lenses of -7 D power. After 24 h, the mRNA expression of selenoprotein increased by about 387%, and the untreated contralateral eye by 786%. Again, the comparisons did not reach significance.
Retinal image degradation in one eye by a diffuser elevated the mRNA level by 261% after 6 h (Dunnetts' test: p<0.001), whereas a binocular exposure caused an upregulation (+425%) after only 2 h (Dunnetts' test: p<0.001). Binocular diffuser treatment (2 h and 4 h) increased selenoprotein mRNA significantly more than monocular treatment (Figure 2, unpaired t-test: 2 h monocular diffuser compared to 2 h binocular diffuser: p=0.0027; 4 h monocular diffuser compared to 4 h binocular diffuser: p=0.0006).
After 24 h, defocus imposed by a +7 D positive lens resulted in a statistically significant upregulation of the prolidase gene by about 75% (Figure 3; ANOVA: TE/CG: p=0.014). Prolidase mRNA levels in the untreated contralateral eye did not change significantly over time. At every time point measured, prolidase mRNA levels were higher in the plus lens treated eyes compared to the contralateral eyes. In the untreated contralateral eye of the group treated with a monocular negative lens, we measured a significant rise (ANOVA UE/CG: p=0.009) of mRNA expression after 24 h (+96%) whereas changes in the minus lens treated eyes did not reach significance (ANOVA TE/CG: p=0.064).
After 4 h of diffuser wear, there was a prominent increase in prolidase mRNA expression levels both with monocular (+142%) and binocular (+106%) treatment. Although the prolidase mRNA level in the untreated contralateral eye was also elevated (+91%), it was still significantly lower than in the deprived eye (paired t-test with Bonferroni correction: p<0.01).
The mRNA level of sequence 3 in animals treated with positive or negative lenses remained unchanged during the treatment, both in the lens treated and open fellow eyes (Figure 4).
However, there was a strong (+228%) increase in mRNA content after 6 h of binocular diffuser wear (ANOVA TE/CG: p=0.028; Dunnetts' test: TE 6 h/CG: p<0.05). In the monocularly treated group, only the untreated fellow eyes displayed a similar increase in expression levels after 6 h (+182%), compared to the untreated control group (ANOVA UE/CG: p<0.001). This change was already significant 4 h after the beginning of the monocular diffuser wear (+122%).
mRNA expression after recovery from blur
Finally, chicks were treated with monocular diffusers for 4 days and gene expression was measured during the recovery period, 4 h after diffuser removal (Table 5). Following the 4 h recovery period, none of the 3 genes under consideration showed a significant change in expression compared to the untreated control group.
Discussion
The housekeeping gene 18S rRNA as an internal control
To control for potential fluctuations in the efficiency of cDNA synthesis that affects the amount of template, it is necessary to co-amplify a reference gene in each sample. This internal control should remain stable in expression during the treatment with diffusers or lenses. In our experiments, 18S rRNA met this criterion. Other authors have reached a similar conclusion, using various experimental designs [9]. However, other researchers preferred other housekeeping genes like β-actin or GAPDH.
Alterations in mRNA expression in the retina after imposed defocus, blur, or recovery from blur
The major finding related to the time course of selenoprotein P expression was a large increase in mRNA level after 24 h of treatment, both with positive and negative lenses. The upregulation was even more pronounced in the untreated fellow eyes. Retinal image degradation imposed by diffusers resulted in a rapid increase of the mRNA levels with binocular treatment, and a slower upregulation with monocular occlusion. Taking together, these results suggest that selenoprotein P mRNA expression changes may not reflect sign of defocus detection but rather be related to blur detection. Previous studies in chickens [7], using differential display, revealed an increase of the amount of mRNA of selenoprotein (although only for a shorter sequence fragment) after a 4 h recovery period subsequent to 4 days of diffuser treatment. In the present study, this effect could not be confirmed by real time PCR. However, it is difficult to compare both experiments. One drawback of the DD study was that no untreated control group was included, whereas this was the case in the present study. Moreover, the number of treated animals was higher in the current real time PCR studies (4-7 compared to 3-4).
We found a significant upregulation of prolidase mRNA levels after 24 h of positive lens treatment but not with negative lenses. In contrast, retinal image degradation induced by diffuser wear caused a prominent increase in prolidase mRNA levels already after 4 h, both with monocular and binocular treatment. The later result was also obtained in the studies using DD. Our findings of an increased retinal prolidase mRNA expression after a 4 h treatment with diffusers and after 24 h of plus lens wear show that the enzyme is not regulated by the sign of defocus.
A sequence that could not be identified in database searches was referred to as sequence 3 in this paper. A fraction of this sequence was also previously investigated in the course of the DD approach. In this study, the mRNA expression of the gene was found to be controlled by the sign of imposed defocus. Positive lens treatment caused an upregulation of the mRNA levels, and diffuser or negative lens treatment resulted in a decline of the mRNA levels. These findings were not reconfirmed in the present study using real time PCR. The level of mRNA expression remained unaffected by imposed defocus, no matter its sign. However, in the previous study, retinal image degradation by diffusers caused a prominent increase after 6 h of binocular diffuser wear. Although both diffuser and minus lens wear induce enhanced eye growth and lead to myopia development, they are in some ways different. Diffuser wear represents an open loop condition because the retinal image does not improve upon enhanced growth, whereas minus lens treatment represents a closed loop condition, in which the eye grows as long as the image gets sharp again. It is therefore quite possible that differences in signal transduction pathways exist as shown here for changes in mRNA levels of sequence 3 solely after diffuser wear. The lack of information on the identity of this gene makes it difficult to suggest a possible function in postnatal eye growth control. However, the current information may still be of interest since the expansion of the chicken data bases will permit its identification soon, and it is likely that the gene product is involved in retinal blur detection.
Previously, Winawer and Zhu investigated the influence of different signs of imposed defocus on the speed of the emmetropization process in the chickens [10,11]. They found that brief periods of myopic defocus, induced by positive lens wear, were sufficient to prevent myopia that would be induced by wearing negative lenses for the rest of the day. According to their experiments, myopic defocus creates a faster and more powerful signal for emmetropization than hyperopic defocus. The current results from the three genes are probably no candidates for such asymmetrical signals because there was not even a significant difference in the changes induced by defocus of either sign, at least not for treatment periods of up to 24 h.
Contralateral eye effect
We found a striking co-regulation of mRNA expression in the contralateral untreated eye. In some cases (e.g., 24 h of negative lens treatment/selenoprotein), the effect on mRNA expression changes was even statistically significantly higher in the untreated eyes. Comparing selenoprotein and sequence 3 gene expression changes in monocularly and binocularly deprived eyes, it became obvious that binocular diffuser application results in faster and more prominent mRNA expression changes compared with monocular diffuser wear. A significant difference between treated and contralateral eyes could only be detected after a 4 h period of diffuser wear, when the amount of prolidase mRNA was significantly higher in the occluded compared to the fellow eye. The phenomenon of co-regulation was earlier described for both the ZENK mRNA and the ZENK protein [2,3,12]. Moreover, Siegwart et al. [5] showed a strong contralateral regulation of mRNA levels in tree shrews. The nature of the interocular connection remains obscure. Both humoral factors and neuronal pathways could be responsible. However, it is puzzling that the actual changes in eye growth are confined to the treated eyes, despite the joint alterations in gene transcription in the fellow eyes. These observations emphasize that it is necessary to have a completely untreated control group available.
The potential relevance of the investigated genes in the visual control of eye growth
The current study describes, for the first time, that selenoprotein P mRNA is expressed in the chicken retina. Coca-Prados et al. [13,14] described the transcript in the human ciliary epithelium, and others described it in the lymphatic endothelium of the trabecular meshwork in humans [15]. Besides the plasma, it is also found in many human tissues like heart, liver, and lung, and in rat kidney, testis, liver, and lung [16]. The role of selenoprotein P in vivo is unknown, even though there is convincing evidence for its function as an antioxidant [17,18], as a selenium transporter [19,20], and as a cell survival promoting factor in the primary culture of neurons [21]. Very recently, Hirashima et al. [22] identified selenoprotein P fragments as a cell death inhibitory factor in cell culture. Given that retinal image degradation evoked by diffuser wear induces growth processes in scleral tissue and that this is accompanied by an upregulation of retinal selenoprotein mRNA, one could consider selenoprotein P mRNA (or parts of it) as a potential go-signal in eye growth. According to the findings from the literature, it seems to be a disinhibitory rather than a growth promoting signal. However, the transcription of selenoprotein P is not regulated by the sign of defocus, and mRNA levels increase after 24 h both with myopic and hyperopic defocus. For future experiments it would be interesting to see if there are also changes in selenoprotein P mRNA in the sclera, where the actual growth processes occur.
Prolidase, also called peptidase D (EC 3.4.13.9), imidodipeptidase, proline dipeptidase or aminoacyl-L-proline hydrolase, is a fairly well described enzyme of the collagen metabolism in humans [23,24]. It has been found in the tissues of various species, like in porcine pancreas, in guinea pig lung and kidney, and in rat kidney, liver, intestine, erythrocytes, and human skin fibroblasts [25-28]. A clinical disorder of prolidase deficiency is associated with pathologic myopia [29]. On the biochemical side, there is evidence that prolidase may be a limiting factor in the regulation of the collagen biosynthesis [30]. Due to its cytosolic exopeptidase activity, it causes an extracellular accumulation of C-terminal proline and hydroxyproline. At present, it is unknown whether the changes in prolidase mRNA levels that we measured in the retina can control the growth of the sclera. Retinal prolidase expression and the resulting accumulation of proline and hydroxyproline could affect scleral growth either directly, or indirectly. In the first case, the modified amino acids would have to be transported to the sclera through the RPE and choroid. In the second case, the vision induced changes in prolidase expression in the retina would have to alter the signaling cascades which, in turn, would trigger scleral growth.
In conclusion, we were able to detect and measure three out of six retinal sequences that were differentially expressed after hyperopic or myopic defocus, image degradation by diffusers, or during recovery. Selenoprotein P, prolidase, and the third, as yet unidentified sequence 3, were quantified as a function of time during various treatment conditions. Even though we found fast changes in gene expression within a few hours, none of the genes showed a change in transcription that was correlated with the sign of defocus. The data rather suggest that prolidase, selenoprotein P, and sequence 3 might interact with other factors which control scleral growth. Further investigations are necessary to clarify their functions.
Acknowledgements
This study was supported by the Waldtraut und Sieglinde Hildebrandt-Stiftung to SO, the Max-Planck Award to FS, and the German Research Council (SFB 430, TP C1). We thank H. P. Wendel for the access to his iCycler to perform the real time PCR.
References
1. Wallman J. Retinal control of eye growth and refraction. Prog Retinal Res 1993; 12:133-153.
2. Bitzer M, Schaeffel F. Defocus-induced changes in ZENK expression
in the chicken retina. Invest Ophthalmol Vis Sci 2002; 43:246-52.
![]()
3. Fischer AJ, McGuire JJ, Schaeffel F, Stell WK. Light- and
focus-dependent expression of the transcription factor ZENK in the chick
retina. Nat Neurosci 1999; 2:706-12. ![]()
4. Feldkaemper MP, Schaeffel F. Evidence for a potential role of
glucagon during eye growth regulation in chicks. Vis Neurosci 2002;
19:755-66. ![]()
5. Siegwart JT Jr, Norton TT. The time course of changes in mRNA
levels in tree shrew sclera during induced myopia and recovery. Invest
Ophthalmol Vis Sci 2002; 43:2067-75. ![]()
6. Ishibashi K, Fujii S, Escano MF, Sekiya Y, Yamamoto M.
Up-regulation of crystallin mRNAs in form-deprived chick eyes. Exp Eye
Res 2000; 70:153-8. ![]()
7. Feldkaemper MP, Wang HY, Schaeffel F. Changes in retinal and
choroidal gene expression during development of refractive errors in
chicks. Invest Ophthalmol Vis Sci 2000; 41:1623-8. ![]()
8. Simon P. Q-Gene: processing quantitative real-time RT-PCR data.
Bioinformatics 2003; 19:1439-40. ![]()
9. Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths
LR. Beta-actin--an unsuitable internal control for RT-PCR. Mol Cell
Probes 2001; 15:307-11. ![]()
10. Winawer J, Wallman J. Temporal constraints on lens compensation
in chicks. Vision Res 2002; 42:2651-68. ![]()
11. Zhu X, Winawer JA, Wallman J. Potency of myopic defocus in
spectacle lens compensation. Invest Ophthalmol Vis Sci 2003; 44:2818-27.
![]()
12. Simon P, Schaeffel F. Temporal sequence of ZENK, RALDH-2 and TGFs-2 expression following hyperopic defocus. 9th International Conference on Myopia; 2002 November 10-14; Hong Kong, China.
13. Coca-Prados M, Escribano J, Ortego J. Differential gene
expression in the human ciliary epithelium. Prog Retin Eye Res 1999;
18:403-29. ![]()
14. Escribano J, Ortego J, Coca-Prados M. Isolation and
characterization of cell-specific cDNA clones from a subtractive library
of the ocular ciliary body of a single normal human donor: transcription
and synthesis of plasma proteins. J Biochem (Tokyo) 1995; 118:921-31.
![]()
15. Tomarev SI, Wistow G, Raymond V, Dubois S, Malyukova I. Gene
expression profile of the human trabecular meshwork: NEIBank sequence
tag analysis. Invest Ophthalmol Vis Sci 2003; 44:2588-96.
![]()
16. Dreher I, Schmutzler C, Jakob F, Kohrle J. Expression of
selenoproteins in various rat and human tissues and cell lines. J Trace
Elem Med Biol 1997; 11:83-91. ![]()
17. Burk RF, Hill KE, Awad JA, Morrow JD, Kato T, Cockell KA, Lyons
PR. Pathogenesis of diquat-induced liver necrosis in selenium-deficient
rats: assessment of the roles of lipid peroxidation and selenoprotein P.
Hepatology 1995; 21:561-9. ![]()
18. Burk RF, Hill KE, Motley AK. Selenoprotein metabolism and
function: evidence for more than one function for selenoprotein P. J
Nutr 2003; 133:1517S-20S. ![]()
19. Burk RF, Hill KE, Read R, Bellew T. Response of rat selenoprotein
P to selenium administration and fate of its selenium. Am J Physiol
1991; 261:E26-30. ![]()
20. Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF,
Burk RF. Deletion of selenoprotein P alters distribution of selenium in
the mouse. J Biol Chem 2003; 278:13640-6. ![]()
21. Yan J, Barrett JN. Purification from bovine serum of a
survival-promoting factor for cultured central neurons and its
identification as selenoprotein-P. J Neurosci 1998; 18:8682-91.
![]()
22. Hirashima M, Naruse T, Maeda H, Nozaki C, Saito Y, Takahashi K.
Identification of selenoprotein P fragments as a cell-death inhibitory
factor. Biol Pharm Bull 2003; 26:794-8. ![]()
23. Endo F, Tanoue A, Nakai H, Hata A, Indo Y, Titani K, Matsuda I.
Primary structure and gene localization of human prolidase. J Biol Chem
1989; 264:4476-81. ![]()
24. Tanoue A, Endo F, Matsuda I. Structural organization of the gene
for human prolidase (peptidase D) and demonstration of a partial gene
deletion in a patient with prolidase deficiency. J Biol Chem 1990;
265:11306-11. ![]()
25. Yoshimoto T, Tsukumo K, Takatsuka N, Tsuru D. An inhibitor for
post-proline cleaving enzyme; distribution and partial purification from
porcine pancreas. J Pharmacobiodyn 1982; 5:734-40. ![]()
26. Denslow ND, Ryan JW, Nguyen HP. Guinea pig membrane-bound
aminopeptidase P is a member of the proline peptidase family. Biochem
Biophys Res Commun 1994; 205:1790-5. ![]()
27. Ganapathy V, Pashley SJ, Roesel RA, Pashley DH, Leibach FH.
Inhibition of rat and human prolidases by captopril. Biochem Pharmacol
1985; 34:1287-91. ![]()
28. Senboshi Y, Oono T, Arata J. Localization of prolidase gene
expression in scar tissue using in situ hybridization. J Dermatol Sci
1996; 12:163-71. ![]()
29. Kiratli H, Satilmis M. Prolidase deficiency associated with
pathologic myopia. Ophthalmic Genet 1998; 19:49-53. ![]()
30. Palka JA. The role of prolidase as an enzyme participating in
the metabolism of collagen. Rocz Akad Med Bialymst 1996; 41:149-60.
![]()