
We editorially reject many papers: Do not let this happen to you.
Absolute “musts” before you submit:
1. Carefully edit your manuscript in American English. Use a copy editor. Write concisely and clearly. Shorter is usually better.
2. Present good data. Insufficient data, small sample groups, or underpowered studies lead to immediate rejection.
3. Conduct sound statistical analyses.
4. Present your data in clearly-labeled, high-resolution figures.
5. Tables must be carefully structured and short. Do not include columns or rows of information irrelevant to the manuscript.
6. Make a significant contribution to vision sciences.
Molecular Vision uses an online manuscript submission system. All new manuscript submissions should use the submission system. Instructions for manuscript preparation and the journal guidelines are detailed below and must be followed.
A major benefit of submitting a manuscript to Molecular Vision is the potential for rapid publication. For this to happen, the following instructions must be carefully followed. Departures from these instructions create more work for the editorial staff and will result in delays. Substantial departures will result in the manuscript being returned to the authors without consideration.
For questions not covered in these instructions, please email the Editors.
Types of Articles:
Molecular Vision encourages the submission of manuscripts on the molecular biology, the cell biology, or the genetics of the visual system (ocular and cortical). Manuscripts should present original, unpublished material not being considered for publication elsewhere and written to be accessible to vision scientists. If accepted, the material (data and text) shall not be published elsewhere without the consent of the Editors and Publisher of Molecular Vision. While there is no limit on the length of a manuscript, manuscripts are expected to be concise.
Following are descriptions of the types of manuscripts Molecular Vision accepts with an outline of the required elements for each manuscript type. Detailed descriptions of the elements of a manuscript are given in the section on "Preparation of manuscripts."
Research Article:
A detailed description of original, unpublished work covering a positive or negative result of significance. Reports of negative results must be investigations that other investigators would be likely to pursue in the absence of the report.
Molecular Vision does not publish simple reports of sequence data or identification of polymorphisms. Reports of new sequence (nucleotide or protein) may accompany related biological data. Simple sequencing data should be submitted to GenBank. New SNPs should be reported to appropriate sites such as http://www.ncbi.nlm.nih.gov/SNP/.
A research article must include the following sections: Title Page, Structured Abstract, Introduction, Methods, Results, Discussion, Acknowledgments, and References. Figures, Tables, Appendices, and Supplements should be included as appropriate.
Technical Brief:
This is a short account detailing a novel method or unique use of current technology that of itself, regardless of the experimental question studied, represents a significant addition to scientific enquiry.
A technical brief must include the following sections: Title Page, Structured Abstract, Introduction, Methods, Results, Discussion, Acknowledgments, and References. Figures, Tables, Appendices, and Supplements should be included as appropriate.
Review:
The Editors solicit suggestions for comprehensive articles reviewing the current status of a particular field or topic. The Editors should be contacted in advance with topics so that appropriateness for publication is confirmed. Appropriateness will be determined by the importance of the topic, lack of existing reviews on topic, and timeliness of request. Reviews of appropriate topics are still subject to peer review; a determination that a topic is acceptable does not guarantee acceptance of a manuscript.
A review must include the following sections: Title Page, Descriptive Abstract, Introduction, Discussion, Acknowledgments, and References. If Results are to be presented, the manuscript should include both a Methods and a Results section. Figures, Tables, Appendices, and Supplements should be included as appropriate.
Preparation of manuscripts:
The sections of the manuscript (Abstract, Introduction, Methods, etc.) should be clearly labelled with the name of the section on a line by itself. Figure captions and Tables should follow the References section. Each paragraph should be preceded and followed by a blank line. The Editors recommend that authors read the Molecular Vision Style Guide before preparing a manuscript for submission.
If any figures, graphs, tables, or data previously published are used, written permission of the publisher of the previous work must be presented. This includes works by the authors of the current submission. Obtaining this permission is the sole responsibility of the author.
Manuscript information
All manuscripts should begin with the title of the manuscript, the names of the authors, the authors' institutional affiliations, and contact information for the corresponding author. Author names should be in order of first name, middle initial, and last name. After each author name, an affiliation needs to be cited in superscript numbers. Each affiliation should start with superscript number and each department at the same University and Institute needs to have a separate affiliation number
We allow only one corresponding author. If the authors wish to have a co-corresponding author, we acknowledge the co-corresponding author in Acknowledgments section and provide the email addresses of both corresponding authors.
Abstract
The abstract should concisely (in fewer than 4000 characters) summarize the work presented. Manuscripts submitted as a Research Article must have a structured abstract consisting of four subsections: Purpose, Methods, Results, and Conclusions. Other types of manuscripts must include a descriptive abstract that details the topics covered in the manuscript.
Introduction
A succinct introduction without subheadings should describe the purpose or goals that led to the production of the manuscript. This should include a concise review of relevant literature.
Methods
This section should detail everything that would be required to replicate the work presented. Non-proprietary names should be used whenever possible. Where relevant, the name of the supplier of items used in the investigation should be identified. Suppliers should be identified by their full company name and location (city, state/country); if the company maintains an online presence, a URL may also be given. The supplier name and location (city, state/country) should be cited in full on first mention and after that, it shall be cited only by company name.
Studies involving animals should include a statement that animal care guidelines comparable those published by the Institute for Laboratory Animal Research (Guide for the Care and Use of Laboratory Animals) or the US Public Health Service (Public Health Service Policy on Humane Care and Use of Laboratory Animals) were followed.
Studies that involve human subjects should indicate that an appropriate institutional review board approved the project. The authors must document that informed consent was obtained. The authors should verify that they adhered to the tenets of the Declaration of Helsinki (JAMA 1997; 277:925-926).
Results
The findings of the investigation should be presented without interpretation or discussion. In short manuscripts where the interpretation of the data is relatively simple, the data may be discussed in a combined Results and Discussion section.
Statistics
Provide sample sizes (N) of each sample group in each experiment. For data presented with statistical analyses, the type of statistical test(s) must be named. The level of significance must be provided. It is the responsibility of the authors to ensure that the assumptions of a given test are met by their data. If this information is not provided, the manuscript will be returned or rejected.
Discussion
An interpretation and commentary on the data (research article) or technique (technical brief) should be presented without speculating beyond the scope of the investigation. For reviews, the discussion is the bulk of the manuscript. It may be sectioned to fit the topic being reviewed and each section may be titled by the author.
Acknowledgments
Authors may briefly mention individuals making significant non-authorship contributions to the manuscript.
If your manuscript was supported in whole or in part by NIH, please include this following text:
This manuscript is the result of funding in whole or in part by the National Institute of Health (NIH) Public Access Policy. Through acceptance of this federal funding, NIH has been given a right to make this manuscript publicly available in PubMed Central upon the Official Date of Publication, as defined by NIH. Supplemental Guidance to the 2024 NIH Public Access Policy: Government Use License and Rights
Funding support for the work presented should be detailed. Specific grant numbers should be provided.
Authors shall disclose any commercial interests in the subject of the manuscript or in entities discussed in the manuscript.
All prior presentation of the manuscript's data at meetings should be indicated; such presentations should not appear among the manuscripts references.
A co-corresponding author can be acknowledged here, and both corresponding authors email addresses can be provided here.
Citations and References
Citations in the text are noted in appropriate places by numbers in brackets (e.g., [3,5,8-12]).
References must be numbered in the order of appearance in the text of the manuscript. All references cited in the text should be listed in the References with corresponding numbers. References should be numbered sequentially and provided as an Arabic number followed by a period. Please do not use parentheses or brackets after or enclosing the reference number. Here is the correct format:
1. Watson JD, Crick FHC. Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. Nature 1953; 171:737-38.
Below are incorrect formats of references:
(1) Watson JD, ...
[1] Watson JD, ...
1) Watson JD, ...
1 Watson JD, ...
References should follow the Vancouver style as described by the International Committee of Medical Journal Editors (Ann Intern Med 1997; 126:36-47) except for part 33 which is outdated (see Example 4 below). This style orders elements of the source of journal articles from least to most specific and has been adopted by the National Library of Medicine.
References to unpublished work should be made parenthetically in the body of the text and not listed as a citation in the References section. If the data comes from some or all of the authors of this work, you may simply list it as "[data not shown]." If the data was provided by some other party (e.g., "John Smith"), the party that conveyed the information should be named as "(Personal communication, Dr. John Smith, Department, Institution, City, State or province, Country)." If you want to provide a more detailed acknowledgement of a person's contribution, you may add that to the Acknowledgements section. Citations to submitted manuscripts are not allowed because there is no guarantee that the source material will ever exist in the form in which it was cited.
Please list all author names in each reference. Do not use “et al”.
Molecular Vision has compiled a page of citation tools with links to tools that are useful in preparing the References section of a manuscript. Additionally, the page includes excerpts of the most commonly used citation types in the Vancouver Style. Remember that incomplete or inaccurate references are not useful to the reader. Following are a few referencing examples that cover what most Molecular Vision articles require:
1. Watson JD, Crick FHC. Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. Nature 1953; 171:737-38.
2. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Press; 1989.
3. Watson JD. How we did it. In: Pauling LC, Kant E, Nietzsche FW, editors. We may have been wrong, progress in thinking. Vol 29. New York: Putnam; 1995. p. 278-99.
4. Wistow G. Peptide sequences for β-crystallins of a teleost fish. Mol Vis 1995; 1:1 <http://www.molvis.org/molvis/v1/a1/>.
Figures
Figures should be numbered with Arabic numerals according to their sequence of appearance in the text, where they are cited as "Figure 1", "Figure 2", etc. Figure captions should follow the References section in the body of the manuscript. Each figure caption should have a title and body. The text of a caption should be sufficient to explain the figure without referring to the body of the manuscript. Captions for figures presenting data must describe the result presented. Labels and abbreviations must also be explained in the caption. Authors should feel free to use color figures in any way that better communicates their message.
Figures that are composites of multiple images (e.g., gels, micrographs) should contain readily discernable white space between the different images. This also applies to composite images of non-adjacent lanes of the same gel.
Figures by number, title, caption, and image file (tiff or jpg) are submitted separately from main text at the submission site. The main text should contain figure captions as well.
Figures must be high resolution, at least 300 dpi, in a full page size image, or on final size image. All text on an image must be legible (at least 8 points) when reduced to final publication size in the PDF. IE, Consider that a single column is only 3.375 inches wide and make sure your text is clearly legible when reduced to that width.
Please see the following examples of bad images:
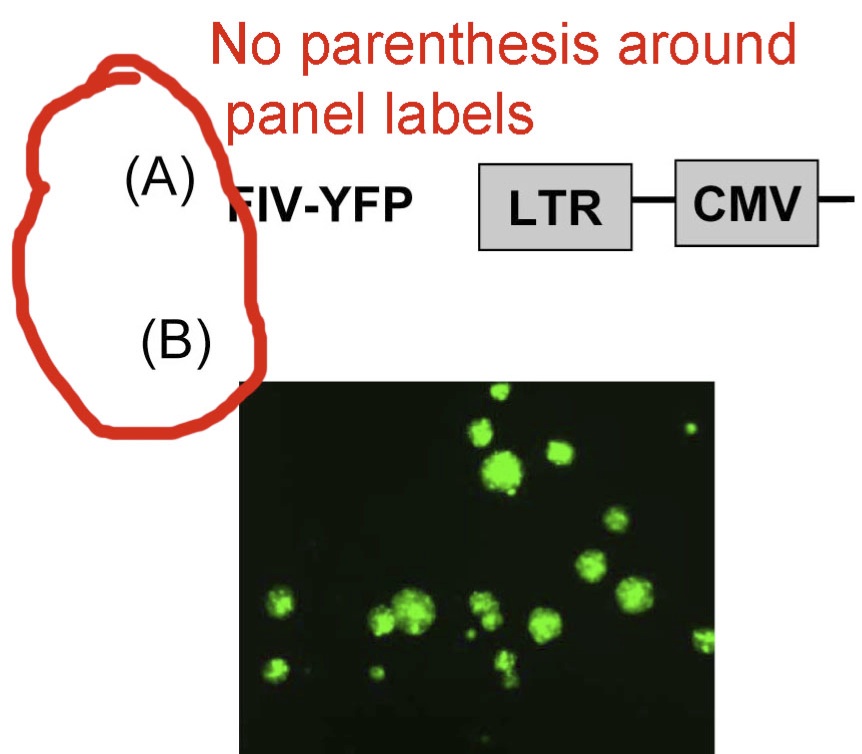
1. Please do not use parentheses in panel labels. Use an uppercase alphabetic character for the panel tag (cf., A, B, C, ….). Do not use numerals. Do not use A1, A2, etc.

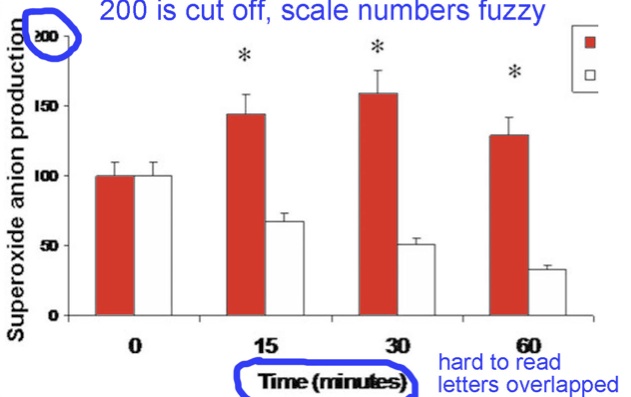
2. Please make sure labels are not cut off, and leave some space between letters. Below, “200” is cut off. The “minutes” label is hard to read.
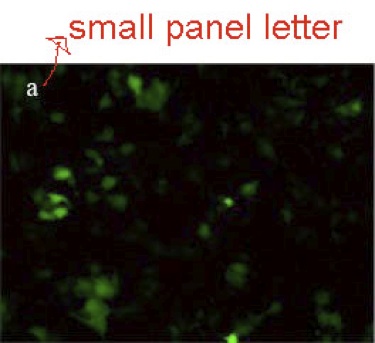
3. Please do not use lowercase panel letters; always use an uppercase letter for each panel tag.
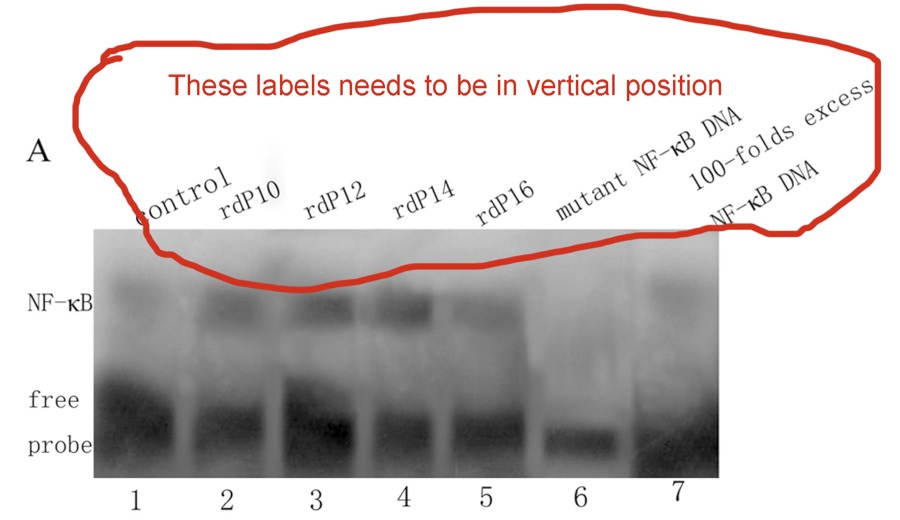
4. In the next panel below, the labeling is legible, but it needs to be in a vertical orientation.
5. In the next panel, panel tags A and B are easy to read but β-actin, 43kd, m-calpain, 97kd, 67kd and 1, 2, 3 are faint and fuzzy. All the labels need to be sharp. Panel tags, A and B, must be in the upper left corner of the image. Note that “kd” is incorrect and it should be “kDa”.

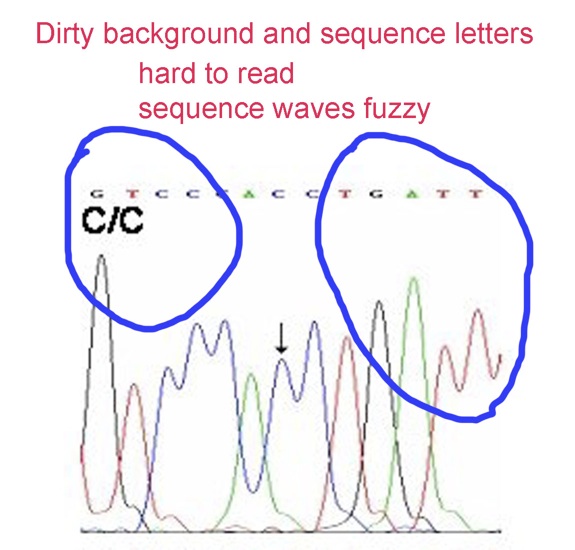
6. In the panel below, especially in images of sequencing chromatograms, uneven background must be corrected. Sequence lettering ought to be replaced with crisp lettering.
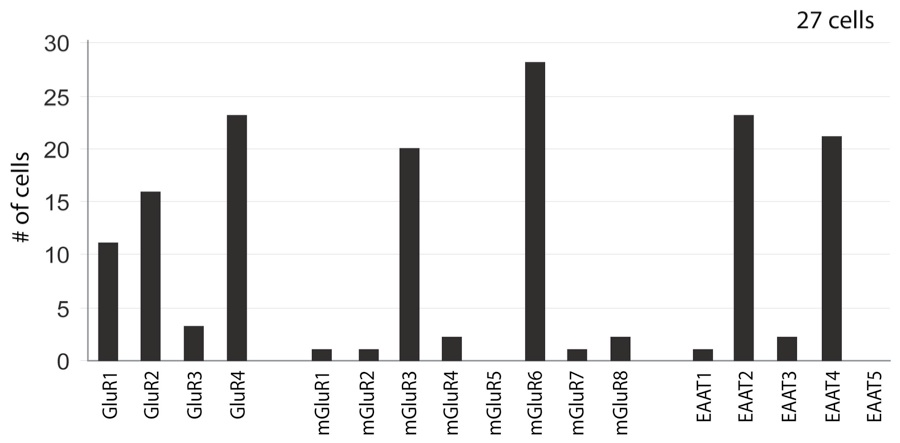
7. In the next panel, the image labels are all sharp. This is the correct display of text on the vertical axis. However, Error Bars are apparently missing in this example.
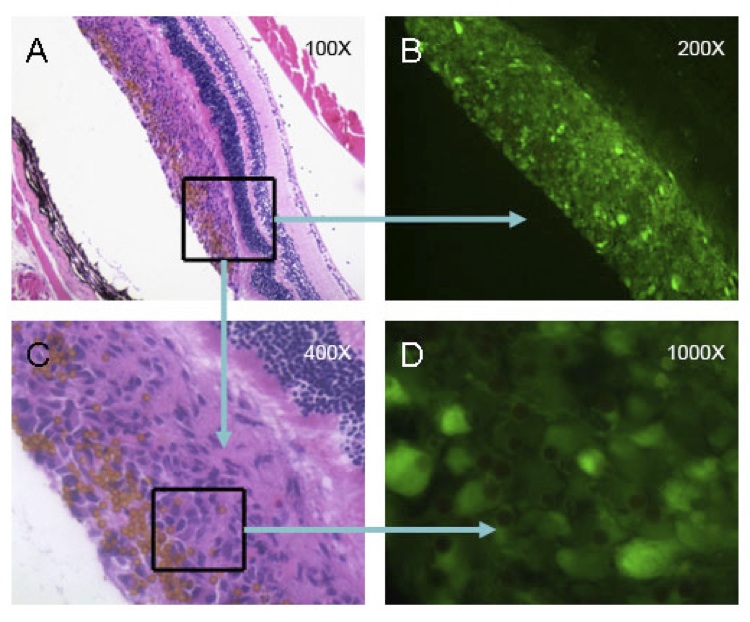
8. In the next image, a figure with multiple panels, the caption should explain each and every panels. All micrographs should contain a scale bar. Please position the scale bar in the lower right corner of the last panel. It is not sufficient to provide the magnification in the caption or on the image.
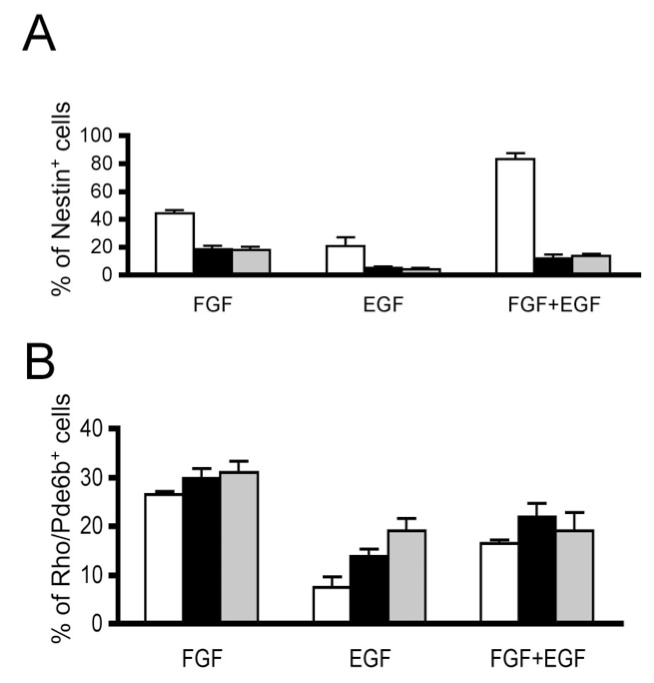
Examples of good figure images:
In the next panels, lines and objects are sharp, and the labels are in the upper left corner. All the other labels are easy to read. Note that Error Bars are shown. These should be defined as either standard deviations or standard error of the mean in the caption, and sample sizes per group should be provided in the caption.
Tables
Tables must be submitted as a Table-formatted Word document. Excel file format is also allowed. Tables should be numbered with Arabic numerals according to their sequence of appearance in the text, where they are cited as "Table 1", "Table 2", etc. All tables should be cited in the text body. Each table should have a title and caption. The text of a caption should be sufficient to explain the table without referring to the body of the manuscript.
Tables that are images are not permitted, and the manuscript will be rejected without review.
Tables must be: 1 page or smaller in a Word or excel file with 1 inch margins on all sides, 12 point font size, Times new roman font style, portrait or landscape and no more than 8 columns wide. Tables longer than one page will automatically be converted to supplementary files and will be shown as appendices.
Appendices
All large tables (over one page in size) and datasets should be provided as appendices and will be regarded as supplementary materials of the publication. All Appendices should be cited in the text body. Please prepare appendices as Word doc tables or as excel files. Other file formats are acceptable but must be approved by the Editors in Chief. These file formats must be compatible with standards set by the National Library of Medicine for publisher submission to PubMed and PubMed Central.
Release of Data
Every manuscript should contain all necessary data for the reader to reach the conclusions reported by the authors. This is much easier to accomplish today because of the public databases available for sequence and structure data. Depositing data in these databases makes it easily available by reference to an identification code that may be cited throughout the literature. Further, it allows for a systematic organization and searching of a class of data.
Authors submitting manuscripts containing newly reported nucleotide or protein sequences must deposit those sequences in the GenBank database. Submissions reporting new gene expression data must submit those data to the Gene Expression Omnibus (GEO). Submissions reporting new three-dimensional structures must submit those structures to the Protein Data Bank. Such submissions should include all structural data supporting the manuscript's conclusions, including any derived atomic coordinates. The accession or identification numbers for these deposits must be provided to Molecular Vision prior to acceptance and must be released (available to the public) prior to publication. It is the authors responsibility to arrange for the release of this information with the relevant database. Any delay in receipt of functioning accession numbers will delay publication. Failure to submit the full datasets will result in rejection of the manuscript. Authors are also required to include accession numbers for any sequences or structures relevant to their manuscript.
Molecular Vision reserves the right at any time to require the submission of other large datasets to major public databases as the editors deem appropriate. These data sets will include metabolomics and protein data sets.
Genome-wide association studies
Molecular Vision encourages authors of genetic association studies to follow prevailing standards (Little et al. PLoS Med 2009; 6: 151-62) for reporting results. Replication of prior studies, whether positive or negative are of value to the community and will receive consideration until the association is clearly established or refuted. Replication studies will receive priority if they extend the work of previous studies and provide a better understanding of the nature of the association. Novel findings in genetic association studies will be given priority if they are replicated in an independent group of subjects. Such replication is important for initial observations and novel findings in replication studies, whether they are derived from genome-wide or candidate gene approaches. Although present standards include guidelines of p <1.0 × 10-5, and sample sizes of at least 250 members per group, these likely will become more strict in the near future. Clearly, sample size and significance depend on the questions being asked and the nuances of the hypotheses, which may require larger sample sizes and stricter levels of significance.
Meta-analyses
A meta-analysis typically is an attempt to identify relationships across several study results, assessing contrast or agreement, and possibly generating new hypotheses whose testing will move the field forward.
For purposes of publishing in Molecular Vision, a meta-analysis must generate new, testable hypotheses.
To quality as a full Research Article a meta-analysis should communicate at least a modest amount of new results (previously unpublished) and should examine aggregated data (e.g., p-value) for each marker (e.g., SNP) for each of numerous SNPs from several prior GWAS studies. The meta-analysis should include clinical criteria for selecting cohorts, ethnicity, gender, genotyping methods, and error rates from the groups that did the initial studies.
A meta-analysis that presents and uses an analytical approach that is new and distinct from previous approaches may be considered a Technical Brief.
A meta-analysis that lacks novel results or findings or analytical approach is a Review Article.
Genetic Analysis
For pedigree analysis and linkage studies, relatively large pedigrees should provide lod scores (>+3.0 or <-2.0) sufficient to accept or exclude linkage to a new locus unless also supported by identification of a high probability mutation in a gene in the included region or other evidence of biological significance. Studies confirming previously published results will be considered as long as they provide additional insight into the existing association or linkage data. This includes identification or refinement of the biological import, the importance of previously identified linkage or associations in specific populations, and the genetic mechanism for population differences.
Polymorphism studies
Polymorphism reports are not generally acceptable unless there are a large number of new polymorphisms reported such that the resource needs explanation in the literature, or the manuscript provides insight into a biological process of general interest. In general, polymorphisms should be deposited in a suitable public database such as dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) prior to publication.
New Mutation Studies
The standards for publication of new mutation studies in "Molecular Vision" have become stricter.
We will still consider manuscripts that describe a gene lesion that causes disease in a new gene, provided that the manuscript data are convincing. Subsequently, for new mutations found in the same gene with the same disease, we expect a larger number of new mutations in each article, due to the increasing ease of detection of causative mutations and a glut of articles. For example, many gene lesions have been identified in rhodopsin that cause retinitis pigmentosa. We will not consider a manuscript on just a single new mutation in rhodospin. Starting now, we would consider two new novel mutations in rhodopsin. We expect that in the near future we will likely expect several new causative mutations per paper. The new standard is three or more new mutations.
Manuscripts that include functional biological
data or experimental data regarding a new mutation will be
given significantly higher priority over those that lack it.
Such manuscripts are not subject to the above rule on the
number of new mutations. To be more competitive for the
limited number of articles that Molecular Vision can publish,
we strongly urge each article to contain supporting
experimental data of this nature.
Molecular Vision policy on use of cell lines: Compulsory authentication
Manuscripts reporting experiments in which immortalized cell lines
are used must include data and documentation that demonstrate that
the actual cells used in the experiments reported in the
manuscript exhibit the correct phenotype and genotype. Authors are
must read the article written by the editors on the authentication
of cell lines (The
Editors. On authentication of cell lines. Mol. Vis. 2013;
19:1848-1851). In brief, authors must demonstrate that the
cells actually used are of the correct species of origin, the
correct sex and genotype, and express genes and gene products that
are specific to the pertinent cell type.
It is expected that the cell line expresses genes necessary for hypothesis testing. This may require specific differentiation. Regardless, correct expression must be documented in the manuscript. Submissions that use primary cell cultures or animals to verify hypotheses tested first in immortalized cell lines will be viewed in general more favorably than work that relies solely on cell lines.
The critical policy, criteria, and instructions for meeting these criteria are quoted here from that article:
"Molecular Vision policy on use of cell lines: Compulsory authentication
The editors of Molecular Vision recognize that the proper use of immortalized cell lines can result in the rapid generation of substantial data in testing many hypotheses. However, due to continual problems of misidentification of cell lines for at least 55 years [24-28], including estimates of mammalian cell misidentification of 15%-35% [27,29,30], the new Molecular Vision policy for reporting data based on immortalized cell lines is:
Manuscripts reporting experiments in which immortalized cell lines are used must include data, documentation, and citations that demonstrate that the actual cells used in the experiments reported in the manuscript exhibit the correct phenotype and genotype. It must be demonstrated that the cells actually used are of the correct species of origin, the correct sex and genotype, and express genes and gene products that are specific to the pertinent cell type. Where possible, phenotype analysis should include the effects of differentiation. Cells used in experiments should be within a few passages of authentication (typically five passages). These standards hold even if the cell lines are considered "established" and were obtained from reputable sources. A statement of cell handling protocol that includes passage information and authentication data, certification documentation, and/or citation of published authentication by the co-authors must be provided in Methods sections of submitted manuscripts. These will be part of the freely-available article.
Meeting these criteria
Authors are encouraged to study the history and consequences of misidentification of cell lines and the array of solutions available to avoid this chronic problem. Several excellent reviews and primary papers are available (e.g., [22,24-28,31]). Even a cursory reading of these sources provides insight into the historical lack of scientific rigor and expensive outcomes in terms of research monies and careers that has plagued the misuse of immortalized cell lines.
The editors of Molecular Vision agree with The International Cell Line Authentication Committee (ICLAC) and the National Institutes of Health (NIH) in their statements of the need for cell line authentication (NOT-OD-08-017) and approaches to accomplish this authentication [25]. The ICLAC provides guidelines for incorporating authentication into good tissue-culture practice (Advice to Scientists: Incorporating Authentication into Everyday Culture Practice). A similar online resource is published by the National Center for Biotechnology Information [30]. Briefly, once a cell line is authenticated, it is expanded to create a "master stock" of cell aliquots for cryopreservation. An aliquot from the master stock is expanded to create a "distribution stock" of cell aliquots. An aliquot from the distribution stock is expanded into aliquots of cells that are used in the actual experiments reported in submitted manuscripts. Thus, by best practices, the cells actually used in experiments should be within five passages of authentication (this is not five passages from the initial establishment of the cell line, which may have occurred many passages earlier). These guidelines must be followed and documented in order for data based on the use of immortalized cell lines to be published in Molecular Vision. Exemptions for incidental use of immortalized cells (e.g., simple expression confirmation assays) might occur following scrutiny by reviewers and editors. The journal’s Instructions to Authors will contain the above policy statement and supporting details, including examples of potential exceptions.
Authentication itself principally involves short-tandem-repeat (STR) profiling using standards and protocols developed by the American National Standards Institute for human cell lines [30,32] and by the National Institute of Standards and Technology for mouse cell lines [33]. The articles reporting the re-characterization of the RGC-5 cell line also provide insight into authentication approaches [20,22,23]. Cell lines also may be authenticated by replicating the experiments published for initial characterization. Authors can either provide data in their submitted manuscripts demonstrating that cell lines were authenticated by these various standards, or they can contract with external services (e.g., ATCC, Promega, Identicell, DSMZ, Genetica, and others not listed) to provide authentication. If contract services are used, documentation from the service must be supplied to Molecular Vision as part of the manuscript submission. Note that only some, but not all, cell lines sold by commercial suppliers are authenticated. It is the authors' responsibility to confirm authentication."
Again, authors will be held to those standards in the editorial (The
Editors. On authentication of cell lines. Mol. Vis. 2013;
19:1848-1851).
Use of the RGC-5 cell line to model retinal ganglion cells
The RGC-5 cell line was originally reported to be derived from postnatal day 1 rat retina cells, expressed markers specific to retinal cells, and was sensitive to trophic factor withdrawal and glutamate toxicity following treatment with differentiation factors (Krishnamoorthy et al., Brain Res Mol Brain Res. 2001;86(1-2):1-12). Several research groups report that even cells obtained from the originating laboratories are not of rat origin and do not express genes and proteins specific to retina or retinal ganglion cells (e.g., Van Bergen et al., IOVS 2009 50:4267-4272, Krishnamoorthy et al., Invest Ophthal Vis Sci. 2013;54:5712-5719)
New manuscripts containing data derived from RGC-5 cells will be editorially rejected without review.
Negative Data
Studies of a negative nature are accepted only if the logic for carrying out the experiment is compelling, it is highly likely that other groups in the field would inevitably perform the same futile studies repeatedly, and such a report would warn them away from wasting time and money. Studies showing an inability to confirm previously reported association or linkage data will be considered depending on comparability to the original study in terms of disease phenotype, population of interest, methodology, and the power of the repeat study to exclude association or linkage (e.g., 95% probability limits for the odds ratio).
Submission of manuscripts:
New manuscript submissions should use our online submission system.
Instructions for warranting author responsibilities and contributions are handled within the online submission system. Each and every author must personally answer these queries. Please assure that all author names and email addresses are correct and current when submitting your manuscript. Each author must have his/her own unique email address. The corresponding author is not authorized to answer on behalf of the other authors. US government employees must answer the same set of questions online as other authors. Errors at this step will greatly delay the completion of the submission process and block the review process.
Author Responsibilities, Licensing, and Copyrights:
Each author shall sign an online statement warranting that he/she authored the work, actively participated in the reported experiments, and he/she approves and agrees with representation of the data in the article. Each author shall warrant that the article is original, is not being considered for publication by another journal, and has not been previously published other than as an abstract.
Each author shall sign an online statement that, if the article is accepted for publication, the article and all supplementary materials will be made freely available online immediately with a Creative Commons Attribution-NonCommercial-NoDerivatives License 3.0, or CC BY-NC-ND 3.0 (see http://creativecommons.org/licenses/by-nc-nd/3.0/ for license terms). The authors retain copyright and grant Molecular Vision an irrevocable, royalty-free, perpetual license to publish and distribute the article, in all formats now known or later developed, and to identify Molecular Vision as the original publisher.
Competing interests
The authors shall declare any competing, commercial, or conflicts of interests, regarding this publication. Each author warrants this in the online submission system.
Resubmission of revised Manuscripts
Revisions to active manuscripts shall be resubmitted through the online manuscript system.
Do not submit a revised manuscript as a new manuscript.
Notices about republication
For noncommercial use, republished material must include a citation to the original work in Molecular Vision and must not be changed from the original. See http://creativecommons.org/licenses/by-nc-nd/3.0/ for license terms.
Articles authored by a student may be reproduced for inclusion in the student's thesis or dissertation, whether in an online repository at the student's institution or through a commercial service used by the student's institution. Such republications must include a citation to the original work in Molecular Vision and must not be changed from the original. They do not require further permissions from Molecular Vision for republication in a thesis or dissertation submitted in partial fulfillment of requirements for an advanced scholarly degree. CC BY-NC-ND 3.0 (see http://creativecommons.org/licenses/by-nc-nd/3.0/ for license terms) applies to derivative works other than the thesis or dissertation.
Commercial publishers shall ask permission from Molecular Vision to republish parts of an article. Specific directions are online at Commercial Permissions. The publisher also shall ask permission of The Authors directly.