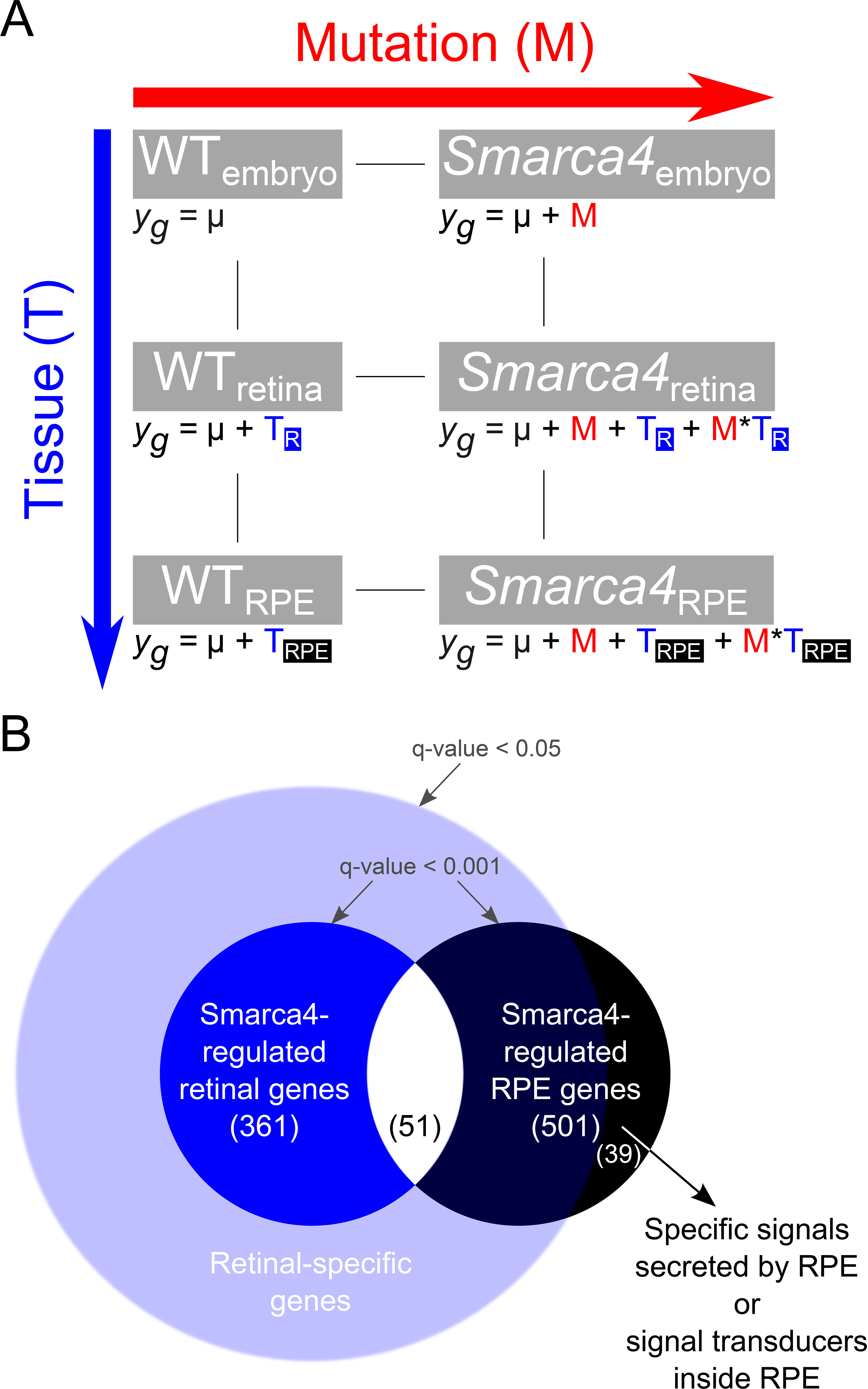
Figure 1. Schematic diagrams of the factorial design and gene selection approach.
A: A 3x2 factorial design used in the expression data analysis. There are two main factors in the model: Tissue (T) and Mutation
(M). The tissue factor T has three levels: whole embryo, retina and RPE, whereas the mutation factor M has two levels: WT
and
smarca4. For each gene
y, its expression value in different experimental conditions was explained by the corresponding equations. The presence of
a factor was represented by a coefficient in the equation. Using the whole family of equations, the statistical modeling explained
the contribution of the individual coefficients in the final expression level.
B: The gene selection was based on statistical testing with the specific combination of the coefficients in the fitted model
for a biological question, i.e. contrast. In this study, we further combined several contrasts to develop rules for selecting
three groups of genes: i) SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member
4 (
smarca4)-regulated RPE genes (black circle;
Table 3 and Appendix 5), ii)
smarca4-regulated retinal genes (dark blue circle;
Table 4 and Appendix 7), and iii)
smarca4-regulated RPE genes that are not differentially expressed in the retina (black circle outside the light blue circle;
Table 5 and Appendix 9). We theorized that genes that were only differentially expressed in RPE but not retina would constitute two
types of genes that may explain the
smarca4 RPE and retinal phenotype. The first type contains signal transducers inside RPE that their dysfunction would cause the
smarca4 RPE defects, while the second type contains signaling molecules that were secreted by RPE that might cause the
smarca4 retinal phenotype. To select this third group of genes, we first obtained a very inclusive group of retinal gene that was
differentially expressed in retina (light blue circle). Then, we excluded these genes from the
smarca4-regulated RPE genes (black circle). Finally, 39 differentially-expressed RPE genes were selected.
 Figure 1 of
Zhang, Mol Vis 2014; 20:56-72.
Figure 1 of
Zhang, Mol Vis 2014; 20:56-72.  Figure 1 of
Zhang, Mol Vis 2014; 20:56-72.
Figure 1 of
Zhang, Mol Vis 2014; 20:56-72.