Received 22 September 2006 | Accepted 5 November 2006 | Published 15 November 2006
Reprint
Received 22 September 2006 | Accepted 5 November 2006 | Published 15 November 2006 |
Download Reprint |
Identification of microRNAs and other small RNAs from the adult newt eye
Evgeny Makarev,1
Jason R. Spence,2
Katia Del Rio-Tsonis,2
Panagiotis A.
Tsonis1
1Department of Biology and Center for Tissue Regeneration and Engineering, University of Dayton, Dayton, OH; 2Department of Zoology, Miami University, Oxford, OH
Correspondence to: Panagiotis A. Tsonis, Department of Biology and Center for Tissue Regeneration and Engineering, University of Dayton, Dayton, OH, 45469-2320; Phone: (937) 229-9257; FAX: (937) 229-2021; email: Panagiotis.Tsonis@notes.udayton.edu
Abstract
Purpose: MicroRNAs (miRNAs) are capable of controlling gene expression by targeting complimentary sequences in many mRNAs. Thus, a small number of miRNAs are capable of regulating expression of many different genes. miRNAs have been found in all animals from Drosophila to human and they are highly conserved. This work was undertaken in order to identify such RNAs in the newt eye.
Methods: Cloning of these RNAs was attempted after isolating and fractionating total RNA from the adult newt eye. A gel slice ranging from about 15 to30 nucleotides in length was cut and the extracted RNA was cloned after several processes involving reverse transcription and linker addition. For expression analysis and verification during the process of lens regeneration we used as a probe mir-124a.
Results: Several microRNAs, piRNAs and other small RNAS were identified. Some of them have eye specific gene targets in other species, but for many a function in the eye remains to be attributed. Expression of miR-124a showed an interesting regulation in the lens regeneration-competent dorsal iris.
Conclusions: The cloned miRNAs and other small RNAs are the first to be reported for this animal and might bear significance in regulating processes that are unique to the newt eye, i.e., regeneration of the lens and retina.
Introduction
MicroRNAs (miRNAs) are short (about 20-23 nucleotides) noncoding RNAs that have the capability of affecting translation of target mRNAs by binding to complementary sequences at the 3' untranslated region, which stops translation [1-3]. Such an action of miRNAs is mediated by the binding and interaction with several key proteins [4]. This is a very interesting secondary level of gene expression control because a miRNA can affect at the same time many different mRNAs which have complementary target sequences [5]. A good example of this is the regulation and clearance of hundreds of maternal mRNAs by mir-430 in zebrafish [6]. As more studies are accumulating, the role of miRNAs becomes more significant during development and differentiation. miRNAs have been isolated in virtually all organisms that have been examined so far and show an astonishing degree of homology (almost 100%) among different animals [1-3,7]. Recently, and as a consequence of the quest for more miRNAs, other small RNAs have also been identified. Some of them show good (but not perfect) homology with known miRNAs, while others are different and vary in size (about 30 nucleotides) [8-10]. It is also becoming accepted that these small RNAs might have significant roles in gene regulation as well. In this paper we report the cloning and identification of several miRNAs and small RNAs from the adult newt eye. These are the first such RNAs identified in this animal and their presence in the eye could bear significance in unique properties of the newt eye, such as tissue regeneration.
Methods
Cloning of miRNAs from adult newt eye; miRNA purification
Total RNA from adult newt eyes was isolated with TRIreagent following the manufacturer's recommendation, with the exception of using 2 volumes of ethanol instead of isopropanol at the precipitation step. For the cloning procedure we used about 70 ug of total RNA. The sample was heated at 65 °C for 5 min, chilled and loaded onto a 15% acrylamide 8 M urea gel (Bio-Rad, Hercules, CA) and was run at 18 volts/cm (150 volts for an 8 cm gel). A ssDNA ladder from IDT was used. The gel was then stained with SybrGold (Invitrogen, Carlsbad, CA) 1:10,000 in 1X TBE for 15-20 min and a slice sized between 15 and 30 nucleotides (nt) was cut. miRNAs were electroeluted with Novagen D-tubes (30 min at 120 V in 1X TBE) following the manufacturer's recommendation. The isolated miRNAs were concentrated using a microcon concentrator unit YM-3 (Millipore, Billerica, MA) following the manufacturer's recommendation and were then washed twice with nuclease free water.
RNA ligation of 3' linker
For this we used the adenylylated oligo "modban" (IDT DNA Technologies, Covalville, IA). It precludes the need to include ATP in the ligation, and hence helps prevent circularization of the RNA. The following 20 μl ligation reaction was set: 13 μl water containing miRNAs purified from total RNA, 2 μl 10X T4 RNA ligase buffer, 2 μl DMSO, 1 μl of 100 μM RNA 3' linker oligonucleotide, "modban" AMP-5'p-5'p/CTG TAG GCA CCA TCA ATdi-deoxyC- 3', 2 μl T4 RNA ligase (20 Units, Amersham, Piscataway, NJ).The reaction was incubated at 37 °C for one h and then it was stopped by the addition of 20 μl of denaturing gel sample buffer (Bio-Rad). The stopped ligation reaction can be stored at -20 °C, or processed immediately by gel electrophoresis (10X T4 RNA ligase buffer: 500 mM Tris-HCl pH 7.5, 100 mM MgCl2, 100 mM DTT, and 600 μg/ml BSA). We then gel purified the product of the ligation reaction by cutting a gel slice between 35 and 45 marker bands. The expected position of ligation product (18 nt Modban plus about 22 nt small RNA) is about 40 nt. This product was electroeluted and concentrated as described above.
RNA ligation of 5' linker
This step is designed to select for Dicer products with a 5' monophosphate [3]. For this the following 20 μl ligation reaction was set: 11 μl 3'ligation product, 2 μl 10X T4 RNA ligase buffer, 2 μl 60 μM ATP, 2 μl DMSO, 1 μl 100 μM RNA 5' linker oligonucleotide,"Nelson's linker" 5'-ATC GTr ArG rGr CrA rCr CrU rGr ArA rA-3', 2 μl T4 RNA ligase (20 Units, Amersham). The reaction was incubated at 37 °C for 1 h and was then stopped by the addition of 20 μl of denaturing gel sample buffer. The product of the ligation reaction was gel purified by cutting a gel portion between 55 and 65 marker bands. The expected size of ligation product (17 nt Nelson linker plus 18 nt Modban plus about 22 nt small RNA) is about 60 nt. miRNAs were electroeluted and concentrated as above.
Reverse transcription
The reverse transcription reaction was set up as follows: 16.5 μl of ligated RNAs in mQH2O and 1 μl 100 μM BanOne: 5'-ATT GAT GGT GCC TAC AG-3' were heated to 80 °C for 2 min and cooled on ice. The following reagents were then added: 6 μl 5X First Strand Buffer (Invitrogen), 1.5 μl 10 mM dNTP's, 3 μl 100 mM DTT, 1 μl RNaseOUT (Invitrogen), and 1 μl SuperScript III RT (200 U/μl). The reaction was heated at 48 °C for 2 min before adding RT. At this point we took out 3 μl for a (-)RT control. The reverse transcription reaction was incubated at 48 °C for 1-1.5 h.
PCR amplification of cDNAs
The following 100 μl PCR reaction (Taq polymerase kit from Fermentas) was set: 2 μl first-strand cDNA, or "minus RT control" (from Section 4), 80 μl water,10 μl 10X PCR Buffer with (NH4)2SO4,1 μl 10 mM dNTP Mix, 4 μl 25 mM Mg2Cl, and 1 μl 100 μM 5' PCR primer, BanTwo: 5'-ATC GTA GGC ACC TGA AA-3',1 μl 100 μM 3' PCR primer, BanOne: 5'-ATT GAT GGT GCC TAC AG-3',1 μl (5 U) Taq polymerase. After brief centrifugation, tubes were placed on a preheated (95 °C) thermal cycler. We used the following PCR program: Step 1; 96 °C 1 min, step 2; 96 °C 10 s, step; 3 50 °C 60 s, step 4; 72 °C 20 s, step 5; 72 °C 3 min. Steps 2-4 were repeated 26 times
The correct size fragment (about 60 nt) was isolated after running the PRC products on a 3-4% GTG Low-Melt agarose gel (1X TAE). One volume (v/w) of 0.4 M NaCl was added and the tube was incubated for 10 min at 70 °C to melt the gel. An equal volume of 70 °C preheated buffered water-saturated phenol (pH 7.8) was then added. After rigorous vortexing the aqueous upper phase was collected and extracted once again with 65 °C phenol. PCR products were concentrated using a microcon concentrator unit YM-5 and washed with TE buffer twice.
Cloning PCR products directly
At this point purified PCR product may be cloned using a TA cloning kit (Promega) according to manufacturer's instructions. The linker oligonucleotide sequences establish unambiguously the original RNA sequence as follows:
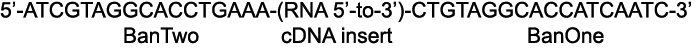
We sequenced several clones to assess the quality of the library before going forward with the concatamerization steps.
Concatamerization of amplified cDNAs
The purified and concentrated PCR product (about 15-20 of 100 μPCR reactions) was dissolved in 500 μl of 1X Ban I NEB buffer 4 and 15 μl Ban I (NEB) was added. The mixture was incubated at 37 °C for 4-5 h. The digestion was tested on a 15% nondenaturing polyacrylamide gel. A fragment of about 40 nt was isolated after running the samples on a 4% GTG Low-Melt agarose gel (1X TAE) as described above. The following ligation reaction (20 μl) was then set: 16μl DNA fragments, 2 μl 10X reaction buffer, 2μl T4 DNA ligase (Ambion), and incubated at 16 °C overnight.
Blunt vector cloning
We then performed end repairing of concatamers (ligation mix from previous section) with End Repair kit (Lucigen, Batavia, IL) following the manufacturer's recommendation. The repairing reaction was resolved on a 2% agarose gel and the 600-1,000 bp material was cut and purified with a standard gel extraction kit (Qiagen, Valencia, CA). The purified concatamers were cloned with ClonSmart kit (Kan LC, Batavia, IL) as described by the manufacturer (Lucigen).
Expression analysis of mir-124a via QPCR
To examine expression of mir-124a during lens regeneration we selected to use QPCR, because we wanted to quantitate its expression during regeneration and because of tissue limitations. RNA from dorsal and ventral irises was isolated using TRIreagent (Molecular Research Center, Cincinnati, OH) following the manufacturer's recommendation, with the exception that 2 volumes of ethanol were used instead of isopropanol at the precipitation step. QPCR was performed using Ambion's mirVanaTM qRT-PCR miRNA detection kit and mirVanaTM qRT-PCR Primer Sets for miRNA-124a. For normalization, Ambion's mirVanaTM qRT-PCR Primer Set for 5S rRNA was used. Real-time PCR was performed with iCycler (BioRad) and SYBR Green I fluorescent dye (Molecular Probes, Carlsbad, CA).
Bioinformatics
All sequences were analyzed using the microRNA registry in the Sanger Database. Due to the lack of newt genome sequences, for target gene identification we analyzed each newt miRNA with the mouse or human genome sequences. From the list of cDNAs that we received we concentrated on the ones with known function in eye tissue differentiation and development.
Results & Discussion
Seventy-two plasmids containing nearly 500 concatamers were sequenced and analyzed for similarity/identity with known miRNAs and other small RNAs available in the miRBase (Sanger database). We identified nine miRNAs, of which four were variables of miR-124 (Table 1). We also identified miR-125b, miR-181b, miR-133a, miR-21, and let-7b. It is interesting to note here that miR-124 has been found to be expressed during mouse eye development, especially in the retina and ciliary body [11]. Also, miR-181 has been shown to be present in the mouse eye as well [12]. let-7b seems to be required for terminal differentiation of cells/tissues at adult stages. Expression of this miRNA could bear significance in the control of cell cycle exit, an important process whose deregulation in the newt is associated with the potential of eye tissues to regenerate [13,14].
In Table 2 we list 19 sequences which are not identical but highly similar to known miRNAs. These miRNAs might be functional since absolute homology with the target mRNA sequence is not required for their effects on protein synthesis. In addition to these sequences, we also cloned four sequences with identity to recently identified piRNAs [9,10]. However, despite the 100% identity, caution should be exercised in accepting two of them as piRNAs because of the homology to tRNA-Met or to 28S rRNA genes (Table 3). piRNAs have been associated with germ cells and their role in the eye is, of course, unknown at the moment. Finally, we identified 42 unknown small RNA sequences that have no similarity to known miRNAs. These sequences were included in the list after we excluded the ones that most likely were the by-product of rRNA or other RNA breakdown (Table 4). However, several sequences showed some low degree of similarity but not enough to be safely excluded due to the lack of newt genome sequences (showed in italics in Table 4).
Our paper describes for the first time the identification of miRNAs and other small RNAs from the adult newt eye. We consider this quite significant, especially in light of the regenerative abilities of adult newt eye tissues such as retina and lens. Regeneration of these tissues is mediated by transdifferentiation of the pigment epithelial cells [15-17]. For such an event to occur the terminally differentiated cells have to rapidly lose their tissue characteristics, re-enter the cell cycle and become a new cell type. It is conceivable that such a transformation requires extensive gene regulation involving hundreds of genes. In this respect miRNAs (each one known to regulate many genes) might play a pivotal role. This is an exciting possibility and the miRNAs and small RNAs that we describe here should become an important tool in examining this. In order to test this possibility, we examined expression of the most commonly cloned miR-124a during lens regeneration. After lentectomy, lens regeneration is achieved via transdifferentiation of the dorsal iris pigment epithelial cells (PECs), but never from the ventral iris. We isolated RNA from intact dorsal and ventral irises as well as from irises 8-days postlentectomy. At this time the dorsal PECs are undergoing the events of dedifferentiation that lead to regeneration of the lens. We evaluated expression of miR-124a via QPCR and the comparisons are shown in Figure 1. As can be seen the most pronounced difference is that miR-124a is expressed at higher levels (nearly 4 fold) in the intact dorsal iris when compared to the regeneration incompetent intact ventral iris, even though the ventral iris does express miR-124a. We have recently shown that the ventral iris is quite active transcriptionally and that regulatory genes are in fact expressed there as well. Thus, induction of regeneration seems to be regulated by levels or expression and not absolute presence or absence of particular factors [17]. At present we do not know the functional role of miR-124a (or of any other microRNA) in lens regeneration but regulation such as the one seen for 124a might indicate fine tuning of gene regulation.
As indicated above, microRNAs can control many genes. Since for the newt there is no genome project data we searched for targets using established databases which utilize mouse or human genomic sequences. Since the newt miR sequences are virtually identical to the corresponding mouse or human ones we reasoned that we can safely deduce target genes. Our search showed that many genes that are pivotal for eye tissue differentiation and development are included as targets of the miRNAs that we cloned (Table 5). For miR-124a, likely targets include microphthalmia-associated transcriptional factor, eyes absent, retinoid X receptor, retinoic acid receptor γ, SOX9, E2F, retinoblastoma-like protein 1, FGFR2, and chordin, all of them known to be involved in eye development. Thus, miRNAs could become very important tools for identifying genes that might regulate eye regeneration as well.
Acknowledgements
Supported by NIH grant EY10540 to P.A.T.
References
1. Carrington JC, Ambros V. Role of microRNAs in plant and animal
development. Science 2003; 301:336-8. ![]()
2. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification
of novel genes coding for small expressed RNAs. Science 2001; 294:853-8.
![]()
3. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny
RNAs with probable regulatory roles in Caenorhabditis elegans. Science
2001; 294:858-62. ![]()
4. Cullen BR. Viruses and microRNAs. Nat Genet 2006; 38:S25-30.
![]()
5. Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle
J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that
some microRNAs downregulate large numbers of target mRNAs. Nature 2005;
433:769-73. ![]()
6. Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue
K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and
clearance of maternal mRNAs. Science 2006; 312:75-9. ![]()
7. Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA
discovery. Nat Genet 2006; 38:S2-7. ![]()
8. Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H.
Stage-specific expression of microRNAs during Xenopus development. FEBS
Lett 2005; 579:318-24. ![]()
9. Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A
germline-specific class of small RNAs binds mammalian Piwi proteins.
Nature 2006; 442:199-202. ![]()
10. Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P,
Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien
M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel
class of small RNAs bind to MILI protein in mouse testes. Nature 2006;
442:203-7. ![]()
11. Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of
mammalian microRNA expression by in situ hybridization with RNA
oligonucleotides. Dev Dyn 2006; 235:2538-48. ![]()
12. Ryan DG, Oliveria-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis 2006; 12:1175-84 <http://www.molvis.org/molvis/v12/a134/>.
13. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A,
Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the
let-7 microRNA family. Cell 2005; 120:635-47. ![]()
14. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC,
Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates
developmental timing in Caenorhabditis elegans. Nature 2000; 403:901-6.
![]()
15. Del Rio-Tsonis K, Tsonis PA. Eye regeneration at the molecular
age. Dev Dyn 2003; 226:211-24. ![]()
16. Tsonis PA, Del Rio-Tsonis K. Lens and retina regeneration:
transdifferentiation, stem cells and clinical applications. Exp Eye Res
2004; 78:161-72. ![]()
17. Grogg MW, Call MK, Okamoto M, Vergara MN, Del Rio-Tsonis K,
Tsonis PA. BMP inhibition-driven regulation of six-3 underlies induction
of newt lens regeneration. Nature 2005; 438:858-62. ![]()
18. Pfaffl MW. A new mathematical model for relative quantification
in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45. ![]()
19. Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free
analysis of quantitative real-time polymerase chain reaction (PCR) data.
Neurosci Lett 2003; 339:62-6. ![]()