![]() Figure 9 of
Cameron, Mol Vis 2005;
11:775-791.
Figure 9 of
Cameron, Mol Vis 2005;
11:775-791.
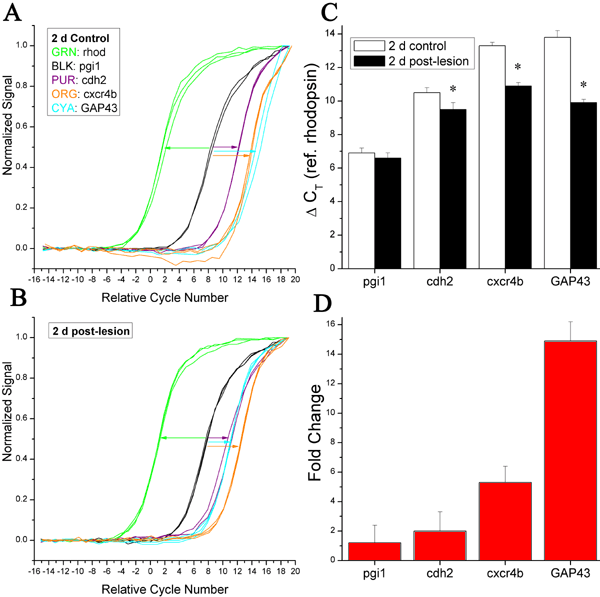
Figure 9. qPCR analysis: 2 d post-lesion retina
Normalized growth curves of single, sequence-confirmed PCR-amplified products are shown for control (A) and 2 d post-lesion retina (B). The amplified products are partial-length sequences (Table 1) encoding rhodopsin (green), pgi1 (black), GAP43 (cyan), cdh2 (purple), and cxcr4b (orange); this color scheme matches that of Figure 6. For ease of comparison the abscissa values are arranged so that 0.0 corresponds to the mean CT value for the rhodopsin (reference) products in that particular condition. Growth curves toward the left of the plot indicate products that are expressed at higher levels than products with growth curves toward the right. The lengths of the colored arrows illustrate approximate differences between the respective CT values, and indicate lesion-induced increases (leftward shifts in the growth curves) in the expression of GAP43, cdh2, and cxcr4b, but not pgi1. C: Measured differences in CT relative to rhodopsin (CT; error bars represent standard deviation). Asterisks indicate statistically significant differences between the control and post-lesion conditions (p<0.05, Student's t-test). D: Inferred linear fold-enhancement of gene expression, determined from the data of panel C. Error bars represent standard deviation.