![]() Figure 4 of
Perkins, Mol Vis 2003;
9:60-73.
Figure 4 of
Perkins, Mol Vis 2003;
9:60-73.
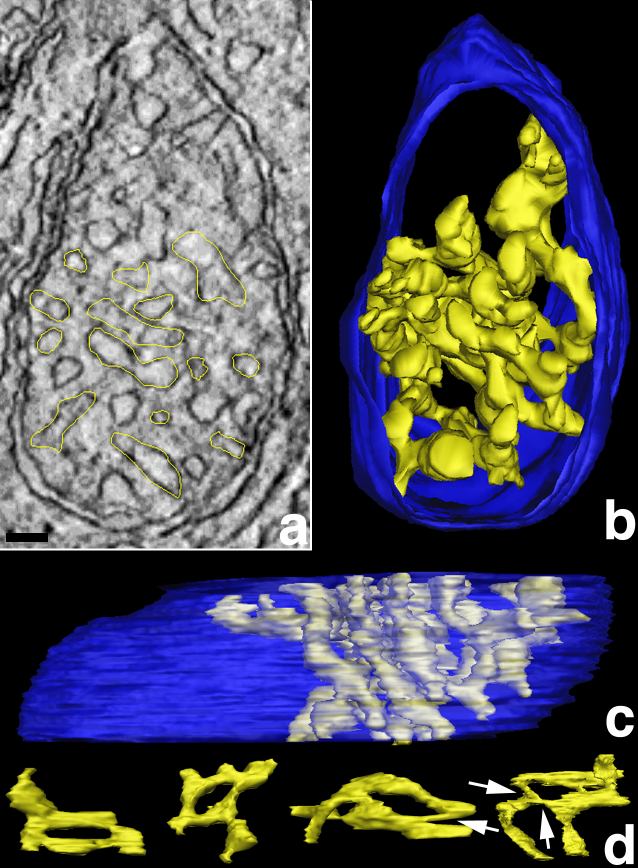
Figure 4. Cone mitochondria have high cristae connectivity
A: Slice through a cone mitochondrion with one traced crista. Connectivity is defined as the sum of continuous connections of cristae segments with each other that form a complete crista. In other words, the connection of well-defined cristae shapes, such as tubes and lamellae, is determined in 3-D. Electron tomography provided this 3-D mapping analysis, which is not feasible with conventional electron microscopy of thin sections. In one 2.2 nm slice of the volume (shown), the yellow tracings highlight all the branches of the largest crista that has the most segments and constrictions of all the cone mitochondrial reconstructions. The connectivity was best mapped after the volume was segmented and surface-rendered. Scale bar represents 50 nm. B and C: Perpendicular views after segmentation of the crista. The crista, traced in A, is shown in yellow. It appears almost maze-like because of its great connectivity of more than 20 segments. The outer membrane is shown in blue. This largest crista measured in cone mitochondria extends through roughly half of the cone mitochondrial volume, has a volume of 110,000 nm3, which represents approximately 9% of the entire mitochondrial volume, and has an inner membrane surface area of 490,000 nm2, which is equivalent to 97% of its total outer membrane surface area. This same connectivity and extent was not observed in rod mitochondria (Figure 5). D: "Loop" connectivity. An unusual crista architecture in cone mitochondria was observed that was rarely noted in rod mitochondria. This architecture is described as loop connectivity because of the connection of crista segments following a closed pathway through the intracristal space back to each other rather than to the inner boundary membrane. Four loops from different cone mitochondria are illustrated. The arrows indicate the narrow connections (constrictions) between loop segments.