![]() Figure 3 of
Gross, Mol Vis 2000;
6:30-39.
Figure 3 of
Gross, Mol Vis 2000;
6:30-39.
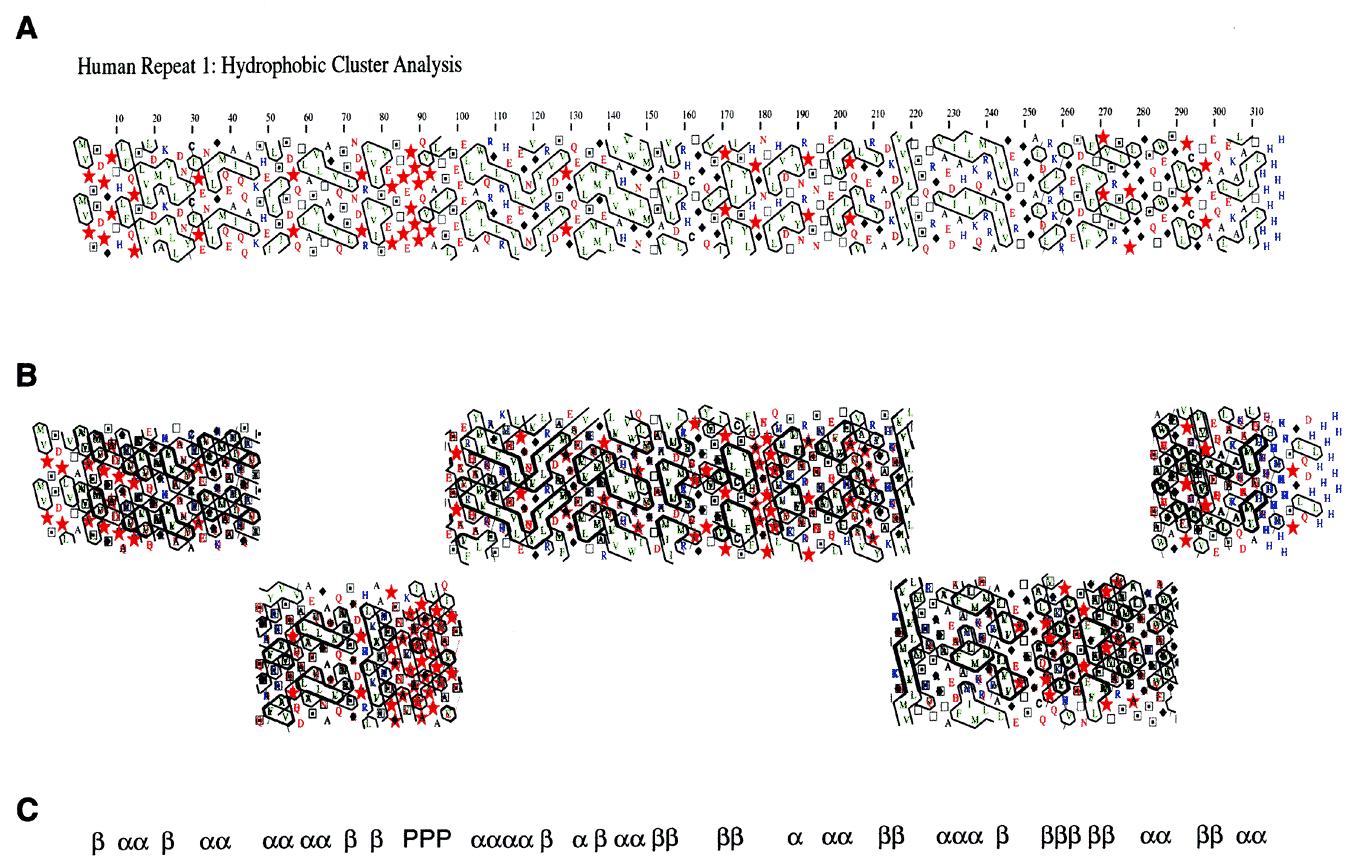
Figure 3. Hydrophobic cluster analysis (HCA) of the four repeats of human IRBP
A primary amino acid sequence is written downward at an angle of about 12.5° from vertical with 7 or 8 amino acids per line, representing about two turns of an a-helix. A second copy of the amino acid sequence is also printed, but it is shifted in phase by 3.5 amino acids. This representation displays amino acids adjacent to each other on the horizontal dimension that might be near each other if they were found in an a-helix. Hydrophobic amino acids are displayed in green and clusters of these amino acids, which include V, I, L, F, M, Y, and W, are boxed by black contour lines. Other amino acids are represented as follows: Red stars, P; black diamonds, G; open boxes, T; boxes with a black dot in the center, S; blue coloring represents basic amino acids (R, K, and H); red letters indicate the acidic amino acids (D, E) and their uncharged counterparts (Q, N). Black amino acids include A and C. The patterns of the contour lines in certain cases are strongly associated with either a-helix or b-strand [16,23]. A shows the hydrophobic cluster analysis of EcR1. Note the clear separation of putative Domain A (amino acids 1-80) from Domain B (amino acids 90-310) by the proline-rich region at about position 85. Positions 100 to 300 correspond to the sequence Hsa_IRBP.1 shown in Figure 2. B shows the conservation of hydrophobic clusters in an alignment of HCAs from all four Repeats (EcR1, EcR2, EcR3, and EcR4). The alignment of the four sequences was done in five blocks to allow four gaps to be introduced at positions likely to contain loops or turns of variable length among the four different sequences. The heavy black lines indicate overlapping contours that are identically positioned among the four sequences. Conserved clusters become obvious and many are associated with one type of secondary structure as predicted in C. The secondary structure assignments were based on the 17 classes identified by Lemesle-Varloot, et al. [16]. For example, the vertical stripes of hydrophobic amino acids at positions 215-220 in EcR1, and well conserved in the other three repeats, was classified as Code 1111, which has a preference ratio of 2.8 to 1, b over a. In the region from 30-70 in Repeat 1 and corresponding regions in the other three repeats, we predict that there may be an a-helix-turn-a-helix structure bounded by a b-strand or extended structure at the N-terminus and another b-strand C-terminal to the last a-helix. In the region from about 160-215 in Repeat 1 and the corresponding regions of the other three repeats there is the same periodicity of 4 prolines: P-hydrophobic cluster-P-hydrophobic cluster-P-hydrophobic cluster-hydrophilic cluster-hydrophobic cluster-P-b-strand. There is a glycine-rich region at about 250 in Repeat 1 and corresponding regions in the other repeats. It is followed by a b-strand at 255-260, a hydrophilic region from 260-265, a glycine-proline rich region from 275-280, hydrophilic regions from 280-290 and 295-305 leading into a possible amphipathic a-helix near the end of the repeat. A possible assignment of the secondary structure based on the conservation of hydrophobic clusters is compiled in C.