![]() Figure 2 of
Gross, Mol Vis 2000;
6:30-39.
Figure 2 of
Gross, Mol Vis 2000;
6:30-39.
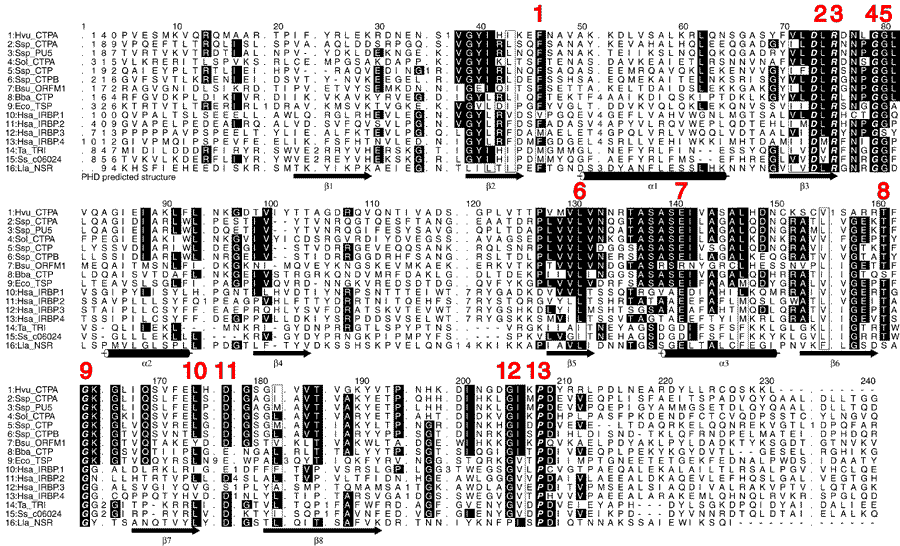
Figure 2. HMM multisequence alignment of the catalytic Domain of Tail Specific Protease and Family members, including IRBP Domain B
Within the 300 amino acid long IRBP repeat unit, there appear to be identifiable structural or folding domains. Domain A in IRBP corresponds to the first 80 amino acids of a repeat. Domain B in IRBP begins near the boundary of Exon 1 and 2 in Repeat 4 and contains amino acids 80-300. Tsp lacks the equivalent eglin c-like structure of Domain A. However, in Tsp and the other carboxy-terminal proteases, they contain a PDZ domain in place of the eglin-like Domain A. Carbohydrate attachment sites are found in the repeat mainly at exon exon boundaries. Predicted secondary structures are labeled on the figure as predicted by PHD. Very highly conserved amino acids are marked by black boxes. All these sequence features are superimposed on an alignment of sixteen different family members. Organisms represented include human, plants, bacteria, cyanobacteria, and archea. Numbers correspond to amino acids changed for mutants 1 through 13 in human Repeat 1 (which are described in a companion paper [12]), sequence 10: Hsa_IRBP.1. Amino acid changes of human Repeat 1 made in the companion paper [12] are shown in red numbers: 1: V116N, 2: L147A, 3: R148D, 4: G152A, 5: G153A, 6: L208A, 7: E218A, 8: T237A, 9: G239T, 10: I249A, 11: E251A, 12: G278A, 13: P281A.
The proteins shown are as follows: Hvu_CTPA is the C-terminal peptidase (Ctp) from the vascular plant Hordeum vulgare (X90929); Ssp_CTPA is the C-terminal processing proteinase precursor from Cyanobacterium synechocystis (A53964); Ssp_PU5 is a hypothetical protein from the petBD operon (P42784); Ssp_CTP (D90906) and Ssp_CTPB (X96490) are other CTPs from that same organism; Sol_CTPA (X90558) is the D1 precursor CTP from the spinach plant (Spinacia oleracea); Bsu_ORFM1 is a CTP from Bacillus subtilis (AF015775 and X98341); Bba_CTP is a CTP from Bartonella bacilliformis (L37094); Eco_TSP (D90827 also known as prc, D00674), is Tail-Specific Protease of Escherichia coli; Hsa_IRBP.1 is Repeat 1 of human IRBP; Hsa_IRBP.2 is Repeat 2 of human IRBP; Hsa_IRBP.2 is Repeat 3 of human IRBP; Hsa_IRBP.4 is Repeat 4 of human IRBP (the accesion number of human IRBP is M22453); Ta_TRI is the Tricorn Protease of archaeon Thermoplasma acidophilium (U72850); Ss_c06024 (Y08256) is a genome sequence from the archaeon Sulfolobus solfataricus. Lla_NSR is the nisin-resistance protein from gram-positive bacterium Lactococcus lactis (U25181).