![]() Figure 2 of
Sergeev, Mol Vis 4:9, 1998.
Figure 2 of
Sergeev, Mol Vis 4:9, 1998.
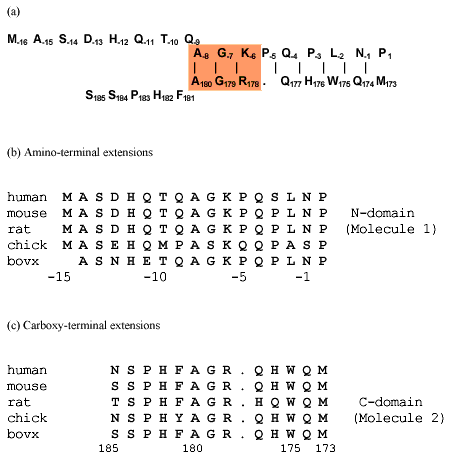
Figure 2. Inverse sequence alignment for mouse ßB2-crystallin
Sequence alignment in opposite orientations of the N- and C-terminal extensions for mouse ßB2-crystallin (a) and for ßB2-crystallin from 5 species (b,c). The shaded area in (a) shows the sequence homology between the N- and C-terminal extensions. The distance between the C[alpha]-atoms of Trp175 and Leu-2 in neighboring arms was 0.517 nm in the first dimer and 0.527 nm in the second dimer of the 1blb file of PDB. The beginning of the alignment was determined for close residues in the ßB2-crystallin structure as shown in Figure 4. The sequence of the N-terminal arm is shown in the usual order from the N- to C-terminus. The sequence of the C-terminal arm starts from the same location in the 3D-structure and is aligned to the N-terminal arm in reverse orientation, from the C- to N-terminus.