![]() Figure 1 of
Sergeev, Mol Vis 4:9, 1998.
Figure 1 of
Sergeev, Mol Vis 4:9, 1998.
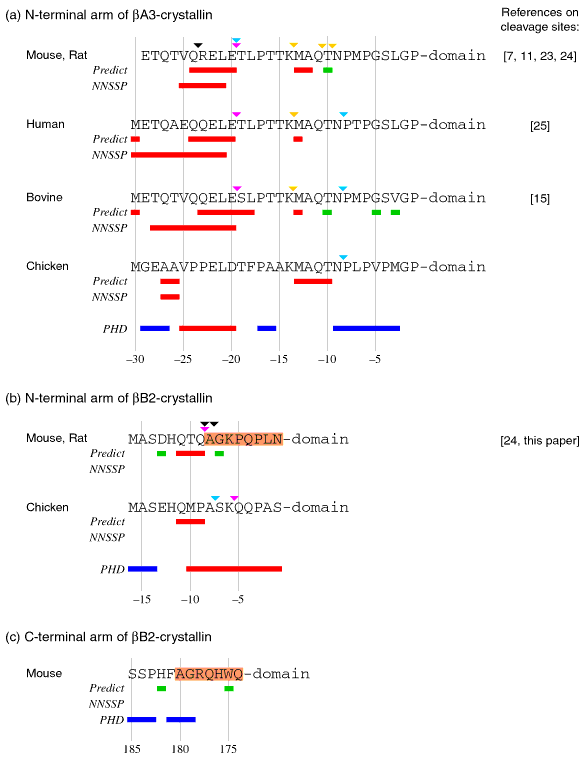
Figure 1. Structural predictions for terminal extensions
The predicted local microdomain structure and partial proteolysis of terminal extensions: (a) N-terminal extensions of ßA3-crystallin, (b) N-terminal extensions of ßB2-crystallin and (c) C-terminal extensions of ßB2-crystallin. Colored bars under each sequence indicate structural predictions based on tripeptide usage (Predict), the NNSSP program (NNSSP), and the PHD method (PHD). Red bars indicate a prediction of a helical region, blue bars indicate a prediction of a coiled region, and green bars indicate a prediction of a turn. Results of the PHD secondary structure prediction are similar for all homologous sequences (more then 40% homology) of each protein. The regions of mouse ßB2-crystallin terminal arm aligned in Figure 2 are shaded. Colored triangles above the sequence indicate cleavage sites. Violet triangles show the major m-calpain cleavage sites, gold triangles show the minor m-calpain cleavage sites, aqua triangles show the naturally occurring cleavage sites, and black triangles show the Sf9 thiol protease cleavage sites with recombinant mouse ßA3- and ßB2-crystallins. The mouse and rat ßA3 and ßB2 sequences were combined because they were identical. However, the Sf9 cleavage sites were determined for recombinant mouse crystallins, while m-calpain and endogenous cleavage sites were determined for rat crystallins. The sites of partial proteolysis were determined as described in the Methods section or were previously published for rat [11,23,24], bovine [15], human [25; David, et al., unpublished data], for chicken (David, unpublished data), and for recombinant mouse ßA3-crystallin by Hope et al. [7].