![]() Figure 6 of
Segovia, Mol Vis 3:9, 1997.
Figure 6 of
Segovia, Mol Vis 3:9, 1997.
Figure 6. Membership of µ-crystallin in an enzyme superfamily
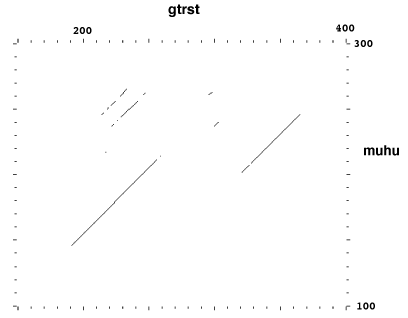
(a) Dot matrix comparison of amino acid sequences for gluTR from Salmonella typhimurium(26) (gtrst) and human µ-crystallin (muhu). Diagonal shows regions of similarity. Comparison drawn using DOTPLOT in the GCG package (27), window size 60, stringency 24.
(b) Sequence comparison of the core conserved regions of gluTR, µ-crystallins and ornithine and lysine cyclodeaminases. Compared regions correspond to residues 122-208 of human and kangaroo µ-crystallin (see Figure 2b). GluTR sequences are: gtrec, Escherichia coli (28); gtrst, S. typhimurium (26); gtrbs, Bacillus subtilis (29); gtrvib, Chlorobium vibrioforme (30). µ-Crystallins are: muhu, human (9 and present study); muka, Grey kangaroo (Macropus fuliginosus) (9). Cyclodeaminase are: ocdach5, Agrobacterium tumefaciens, plasmid Ach5 (11); ocdc58, A.tumefaciens, plasmid C58 (10); lcd, lysine cyclodeaminase of Streptomyces hygroscopicus (20); ocdr, Rhizobium meliloti (31). Regions conserved among members of all three protein families are boxed. Other regions show more local conservation between pairs of families.
