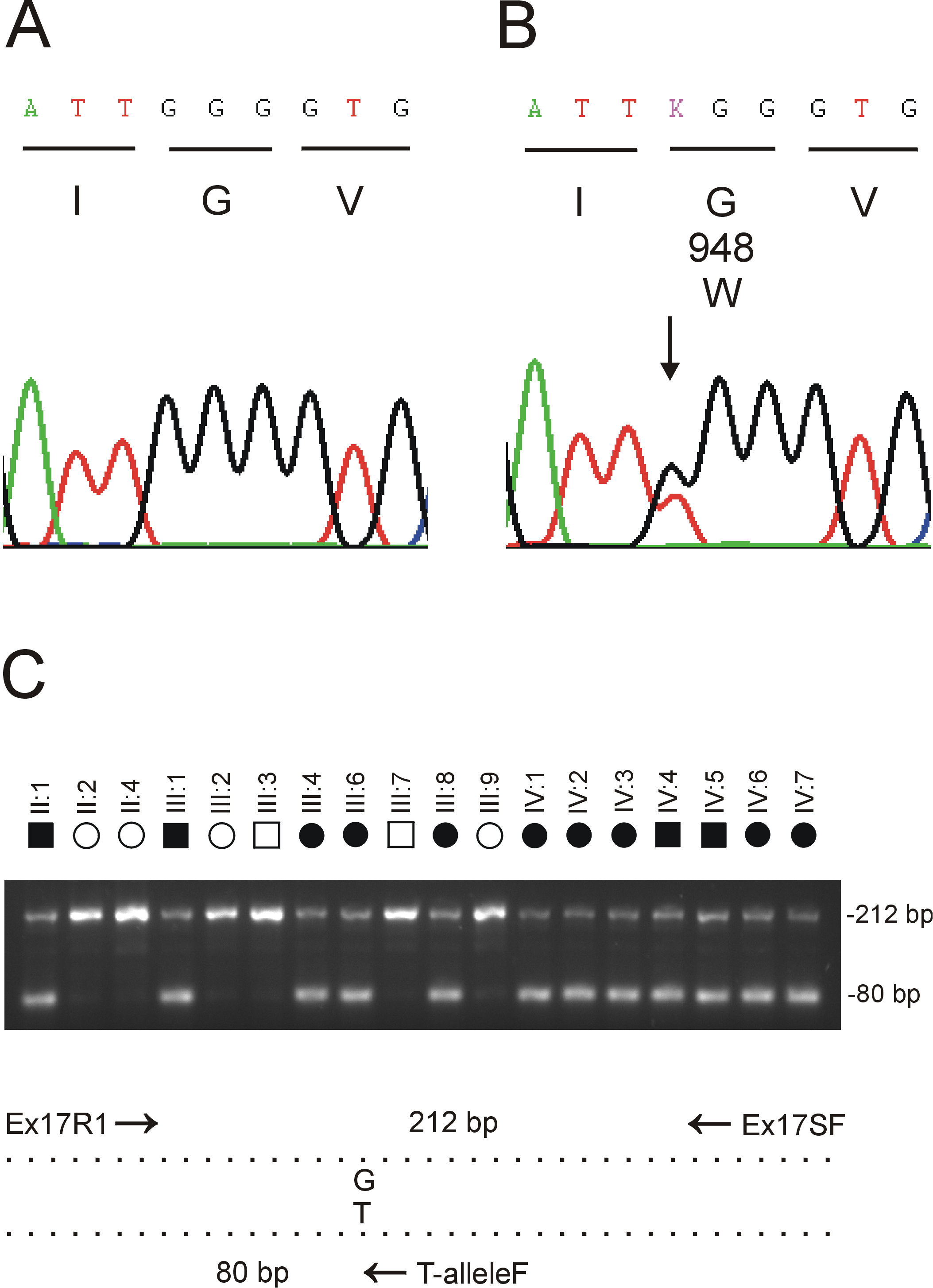
Figure 3. Mutation analysis of
EPHA2
in family Mu.
A: Sequence trace of the wild type allele showing
translation of glycine (G) at codon 948 (GGG).
B: Sequence
trace of the mutant allele showing the heterozygous c.2842G>T
transversion (denoted K by the International Union of Pure and Applied
Chemistry [IUPAC] code) that is predicted to result in the missense
substitution of tryptophan (TGG) for glycine at codon 948 (p.G948W).
C:
Allele-specific PCR analysis using the 3 primers (
Table 1)
indicated by arrows in the schematic diagram; exon-17 was amplified as
above with the sense (anchor) primer located in intron 16 (Ex17R1), the
anti-sense primer located in the 3′-untranslated region (Ex17SF), and
the nested mutant primer specific for the T-allele in codon 948
(T-alleleF). Note that only affected members of family Mu are
heterozygous for the mutant T-allele (80 bp).
 Figure 3 of Shiels, Mol Vis 2008; 14:2042-2055.
Figure 3 of Shiels, Mol Vis 2008; 14:2042-2055.  Figure 3 of Shiels, Mol Vis 2008; 14:2042-2055.
Figure 3 of Shiels, Mol Vis 2008; 14:2042-2055.