![]() Figure 4 of
Yip, Mol Vis 2007;
13:2183-2193.
Figure 4 of
Yip, Mol Vis 2007;
13:2183-2193.
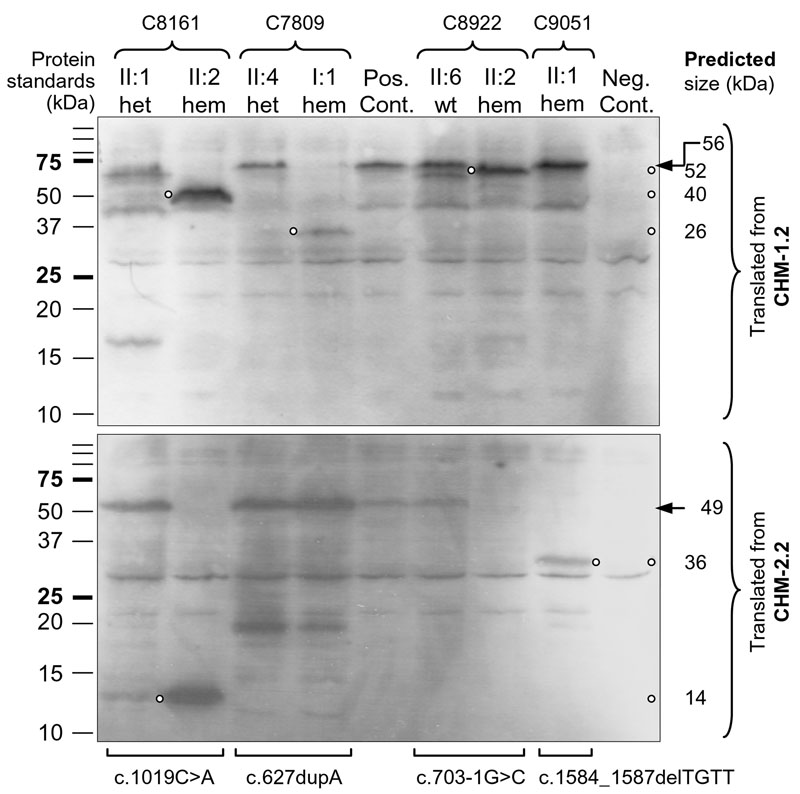
Figure 4. Protein truncation test for CHM mutations
Top gel presents proteins in vitro translated from CHM-1.2 (see Table 2 and Figure 1) and separated in SDS-PAGE, and bottom gel shows those from CHM-2.2. Individuals (II:1, II:2, etc) from different families (C8161, C7809, etc) are listed above the gels and can be identified by referring to the family trees in Figure 2. The corresponding mutations are marked below the bottom gel while the genotypes of the individuals studied are indicated as heterozygous (het), hemizygous (hem), or wildtype (wt; i.e., homozygous normal) above the top gel. Both positive and negative controls (Pos. Cont. and Neg. Cont.) are included in each gel. Precision Plus Dual Color Protein Standards (Bio-Rad) are to the left of the gels while the predicted size of in vitro translated proteins are to the right. Small open circles pinpoint the positions of the truncated proteins while arrows indicate the position of the wildtype proteins. For family C8161, only the results from two representative individuals are shown: II:1 is a heterozygous carrier and II:2 is affected by CHM. Two other carriers (I:2 and II:4), who provided a second sample for testing gave results identical to those of II:1, while another individual with CHM (II:5) also gave the same results as II:2.