![]() Figure 2 of
Lopez-Garrido, Mol Vis 2006;
12:748-755.
Figure 2 of
Lopez-Garrido, Mol Vis 2006;
12:748-755.
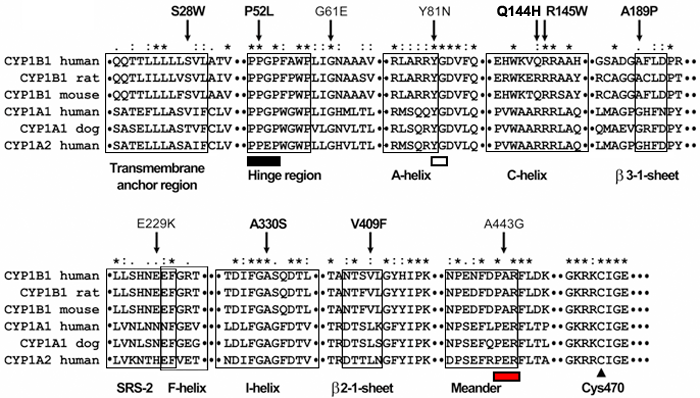
Figure 2.
Multiple amino acid sequence alignment of CYP1B1, CYP1A1, and CYP1A2 from different species. Sequence alignment was generated by ClustalW. Residues affected by mutations are indicated by arrows. Different structural domains of the cytochrome P450 superfamily are boxed. Pro-Pro-Gly-Pro, Pro-X-Arg/His, and X-G motives are indicated by black, white, and red rectangles below the sequences, respectively. The invariant cysteine residue in the heme-binding region is denoted by an arrowhead. Asterisks represent amino acid positions at which all query sequences are identical. Amino acid positions at which all analyzed sequences have amino acids that are chemically similar are denoted by colons (:). One dot denotes amino acid positions with weak chemical similarity. Novel mutations are indicated by bold face type.