![]() Figure 2 of
Mackay, Mol Vis 2004;
10:155-162.
Figure 2 of
Mackay, Mol Vis 2004;
10:155-162.
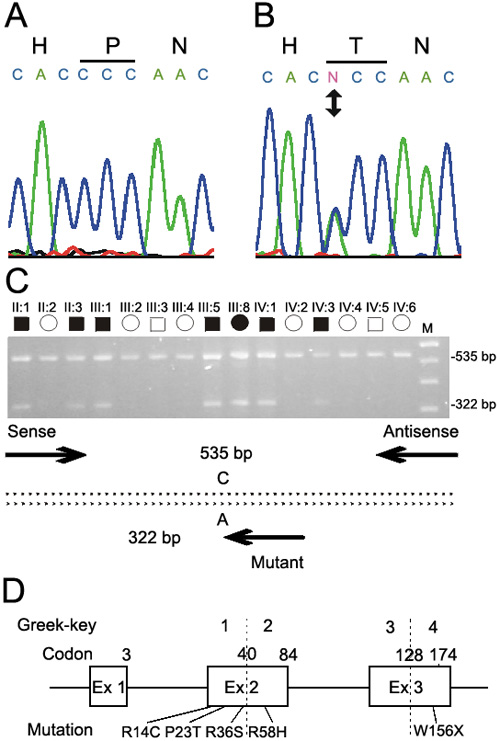
Figure 2. Mutation analysis of CRYGD
Sequence chromatograms of the wild type allele (A) showing translation of proline (CCC) and the mutant allele (B) showing a C->A transversion that substituted threonine (ACC) for proline at amino-acid position 23 (P23T). C: Agarose-gel electrophoresis of allele-specific primer extension products generated using the three PCR primers (Table 1) indicated by arrows in the schematic diagram; the sense anchor primer was located in the 5'-upstream region, the antisense primer was located in intron 2 and the nested mutant primer was specific for the A-allele. Unaffected relatives are homozygous for the wild type C-allele (535 bp), whereas, affected relatives are heterozygous for the mutant A-allele (322 bp). The letter M designates a 100 bp size ladder. D: Exon organization and mutation profile of CRYGD. Codons and corresponding Greek-key protein domains are numbered above each exon. The relative locations of the P23T mutation and four other mutations associated with cataract in humans are indicated. Mutations are numbered according to their amino-acid position in the processed CRYGD protein, which lacks an N-terminal methionine.