![]() Figure 3 of
Hackam, Mol Vis 2003;
9:262-276.
Figure 3 of
Hackam, Mol Vis 2003;
9:262-276.
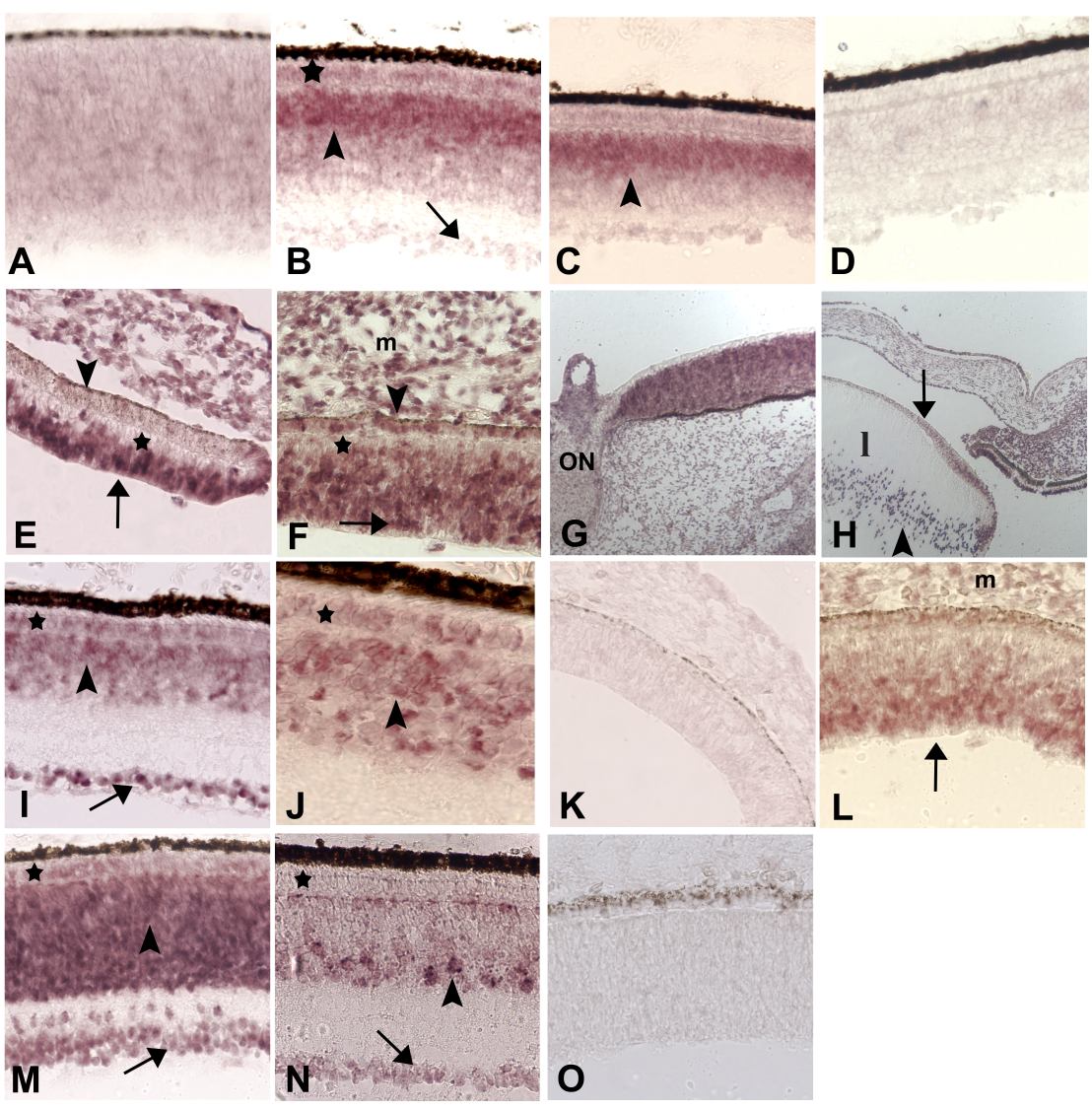
Figure 3. Developmentally-regulated expression patterns of novel chick retina genes
A-D: CRG92, unassigned identity. A: ED 8. Light and diffuse hybridization signals. B: ED 15. Outer part of INL (arrowhead) shows strong signals; ganglion cells (arrow) and the ONL (*) are very light or negative. C: ED 18. Similar pattern as ED 15. E-K: CRG111, homologous to human LATS. E and F: ED 5. Strongly positive retinal neural epithelium is seen towards the vitreal surface at the periphery (E) and fundal region (F, arrows), but not near the RPE (*). The RPE is negative at the periphery (E, arrowhead) but appears positive in the midperiphery (F, arrowhead). Note signal in the periocular mesenchyme (m in panel F). G and H: ED 8. G: Homogeneously positive fundus adjacent to the optic nerve (ON). H: Anterior segment of the eye. Negative anterior epithelium of the lens (l, arrow), and perinuclear signals (arrowhead) in elongating lens fibers. I and J: ED 18. Widespread expression is accompanied by stronger signals in the GCL (I, arrow) than in the INL (arrowhead) and ONL (*). Higher magnification analysis showing heterogenous signals in both layers (panel J); positive cells are indicated (*, arrowhead). L-P: CRG137, homologous to mouse seizure-related gene SEZ-6. L: ED 5. Signal expression throughout the retinal neural epithelium, which is stronger near the vitreal surface (arrow). The mesenchyme (m) is also positive. M: ED 12. Expression in ganglion cells (arrow), the INL (arrowhead) and some cells in the ONL (*). N: ED 18. Restricted expression pattern in mature retinas. Many (but not all) cells are positive in the amacrine layer (arrowhead), accompanied by lightly positive ganglion cells (arrows), and negative photoreceptors (*). D, K, and O: Sense control probes for CRG92, 111 and CRG137, respectively.