![]() Figure 1 of
Hackam, Mol Vis 2003;
9:262-276.
Figure 1 of
Hackam, Mol Vis 2003;
9:262-276.
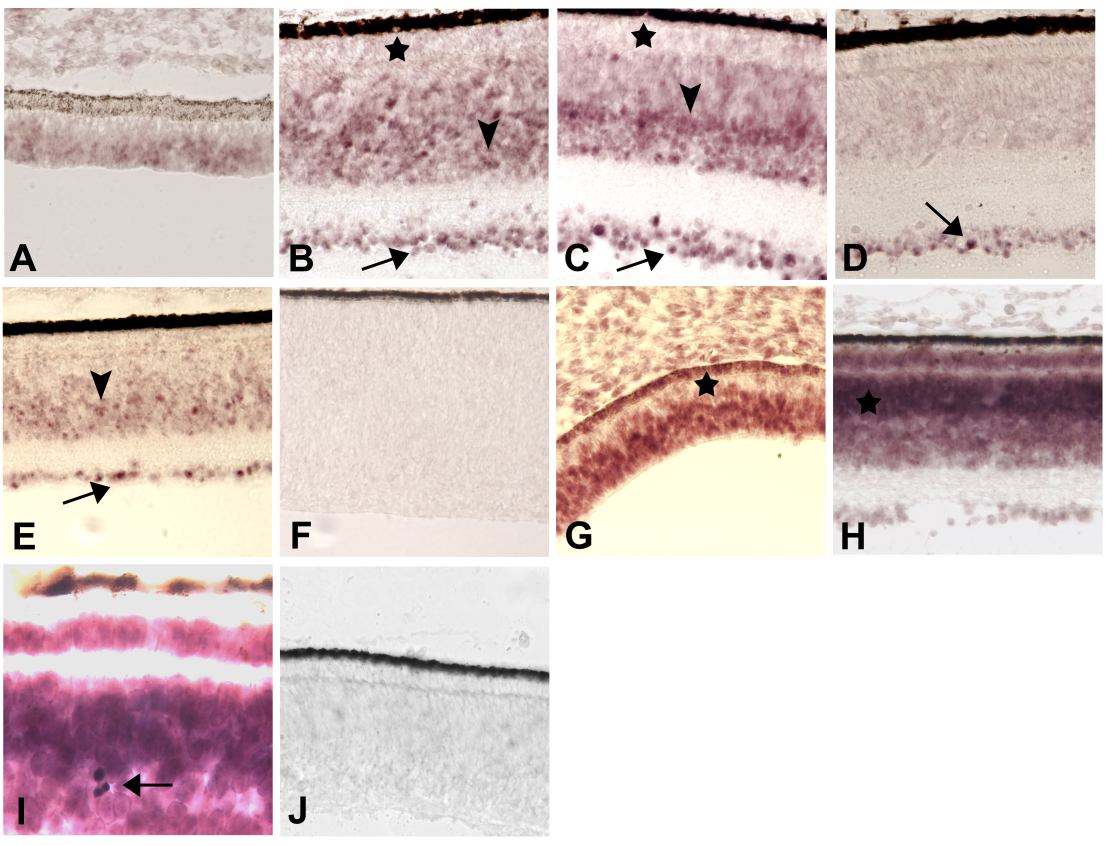
Figure 1. Topographically-specific, developmentally-regulated expression of novel chick retina ESTs
A-F: CRG73, homologous to an EST from a chick eye library. A: ED 5. Hybridization signals are detectable at the retinal periphery but not at the retinal fundus (not shown). B: ED 12. Positive cells are seen in the ganglion layer (arrow) and within the INL (arrowhead). The ONL (asterisk) is largely negative. C: ED 15. Expression is shown in the GCL (arrow) and the ONL (*), and strongly expressing cells in the middle part of the INL (arrowhead). Signals are weaker in the external half of the INL, adjacent to the ONL. D-E: ED 18. Positive cells in the INL at the retina periphery (E, arrowhead) but not at the fundus (D) or the midperiphery (not shown). Ganglion cells are positive throughout the retina (arrows in D and E). G-K: CRG123, homologous to an EST from a normalized chicken pituitary-hypothalamus-pineal cDNA library. G: ED 5. Hybridization signals are strong in the neural retina, except near the RPE (*). H and I: ED 15. Widespread expression in all retinal layers; the outer part of the INL (*) has stronger signals than its inner part. I: Higher magnification showing very dense, strongly positive cells with a circular outline (arrow) whose identity remains undetermined. F and J are sense control sections for CRG73 and CRG123, respectively.