![]() Figure 2 of
Darden, Mol Vis 2003;
9:191-199.
Figure 2 of
Darden, Mol Vis 2003;
9:191-199.
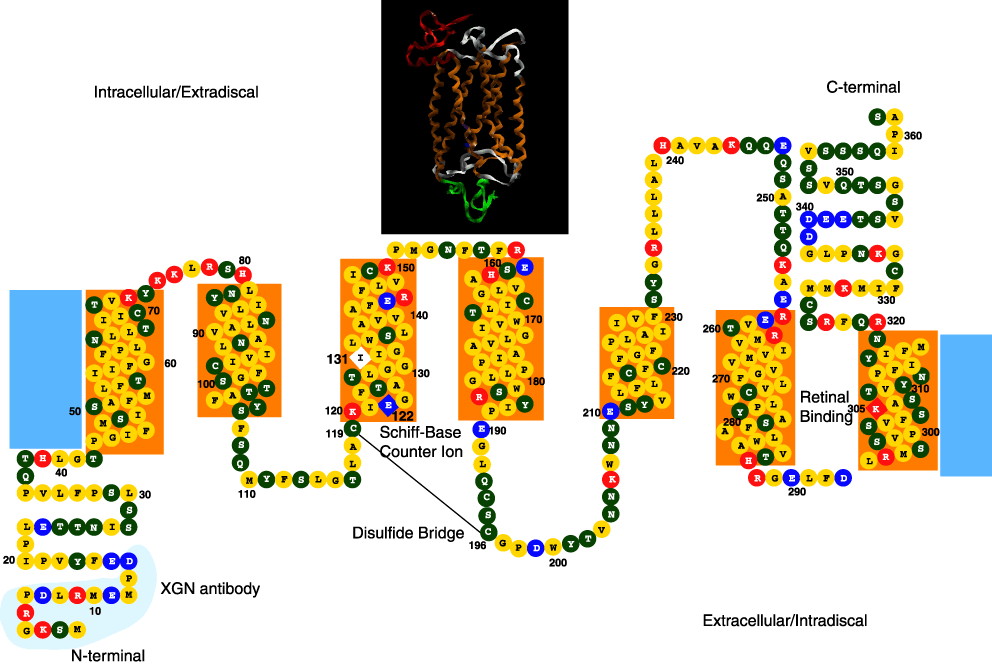
Figure 2. Xenopus SWS2, P434 predicted protein structure
Predicted secondary structure of the SWS2, P434 pigment. The transmembrane domains (boxed) were defined based on Kyte-Doolittle hydropathy plots and the comparison with bovine rhodopsin. The lysine residue at the chromophore attachment site (305), and the Schiff-base counterion glutamic acid residue (122) are indicated by the diamond boxes. The colors of the amino acid beads correspond to the charge of the amino acid: yellow represents the non-polar amino acids; green the polar amino acids; red the basic amino acids; and blue the acidic amino acids. The inserted box shows the predicted three-dimensional ribbon structure based on homology modeling using Modeller with 1HZX as the template. The regions of orange ribbon correlate to the orange transmembrane boxes; the green ribbon to the N-terminus; the red ribbon to the C-terminus; while the white ribbon represents the extracellular and intracellular regions that connect the seven transmembrane helices.