![]() Figure 1 of
Wilson, Mol Vis 2002;
8:259-270.
Figure 1 of
Wilson, Mol Vis 2002;
8:259-270.
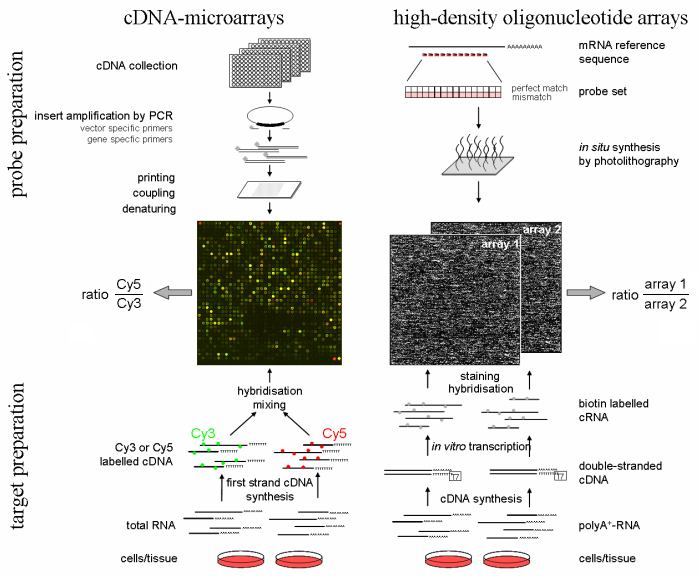
Figure 1. Schematic overview of spotted cDNA microarrays and high-density oligonucleotide arrays
cDNA microarrays: Array preparation: inserts from cDNA collections or libraries are amplified and the PCR products printed at specified sites on glass slides using high-precision arraying robots. These probes are attached by chemical linkers. Target preparation: RNA from 2 different tissues or cell populations is used to synthesize cDNA in the presence of nucleotides labeled with 2 different fluorescent dyes (eg: Cy3 and Cy5). Both samples are mixed in a small volume of hybridization buffer and hybridized to the array, resulting in competitive binding of differentially labeled cDNAs to the corresponding array elements. High resolution confocal fluorescence scanning of the array with two different wavelengths corresponding to the dyes used provides relative signal intensities and ratios of mRNA abundance for the genes represented on the array. High-density oligonucleotide microarrays: Array preparation: sequences of 16-20 short oligonucleotides (typically 25mer) are chosen from the mRNA reference sequence of each gene, often representing the unique part of the transcript. Light-directed, in situ oligonucleotide synthesis is used to generate high-density probe arrays containing over 300,000 individual elements. Target preparation: Total RNA from different tissues or cell populations is used to generate cDNA carrying a transcriptional start site for T7 DNA polymerase. During IVT, biotin-labeled nucleotides are incorporated into the synthesized cRNA molecules which is then fragmented. Each target sample is hybridized to a separate probe array and target binding is detected by staining with a fluorescent dye coupled to streptavidin. Signal intensities of probe array element sets on different arrays are used to calculate relative mRNA abundance for the genes represented on the array.
Modified and reprinted with permission from Nature Cell Biology (Vol. 3, No. 8, pp. E190-E195) Copyright ©2001 Macmillan Publishers Limited.