![]() Figure 6 of
Wistow, Mol Vis 2002;
8:205-220.
Figure 6 of
Wistow, Mol Vis 2002;
8:205-220.
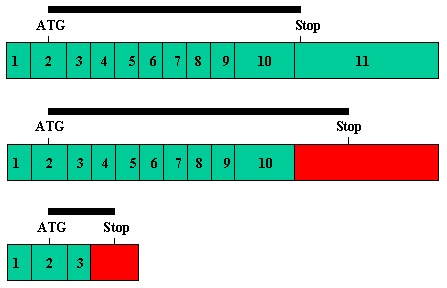
Figure 6. Alternative splicing in Bestrophin
A: Detail of the splice junction skipping that gives rise to a truncated form of Bestrophin. Sequences from Exon 3 of the gene are shown in black, sequences derived from read through into the ORF of Intron 3 are shown in green with the partial predicted amino acid sequence. B: Detail of the splice junction skipping that gives rise to a longer form of Bestrophin. Exon 10 and Intron 10 sequences are in black, Exon 11 sequences are in red. Above shows the expected splice from Exon 10 to 11. Below shows the read through into ORF in Intron 10. C: Compilation of the three Bestrophin protein sequences represented by cDNAs in the cs collection. The sequence of the longest version (from Intron 10 read through) is shown in black and numbered. Sequence derived from the read through into Intron 3 is shown in green. Sequence from the splice to Exon 11 is shown in red. Cysteine (orange) and histidine (blue) residues in the extended ORF form a pattern similar (but not identical) to zinc fingers. D: Cartoons of the three transcript forms of Bestrophin found in the cs collection. Numbered boxes in green show canonical exons for Bestrophin. Red boxes show exon extensions into intron sequence. The positions of start (ATG) and stop codons are indicated. The black bar shows the relative length of ORF for each transcript.
A
Q Q L M F E K L T L Y C D S Y I Q L I P I S ACAACAGCTGATGTTTGAGAAACTGACTCTGTATTGCGACAGCTACATCCAGCTCATCCCCATTTCCT Exon 3 Intron 3 F V L G E F P L L A V P G P C G R P G S R Q A TCGTGCTGGGTGAGTTCCCCCTTCTGGCTGTTCCGGGTCCCTGTGGCCGCCCAGGCTCCAGACAGGCC R G G S R G A A A R G W G G G... AGGGGAGGATCACGAGGAGCTGCGGCAAGGGGCTGGGGAGGGGG... |
B
Exon 10 Exon 11
W A L E N R D E A H S *
TGGGCCTTGGAAAACAGGGATGAAGCACATTCCTAACCTGCTTCCTAATGGGGATGCTTCGCCAGCCAGG
TGGGCCTTGGAAAACAGGTCTGTCCTCCACCTGAACCAGGGGCACTGCATTGCCCTGTGCCCCACCCCAG
W A L E N R S V L H L N Q G H C I A L C P T P...
Exon 10 Intron 10
|
C
MTITYTSQVANARLGSFSRLLLCWRGSIYKLLYGEFLIFLLCYYIIRFIYRLALTEEQQL 60
MFEKLTLYCDSYIQLIPISFVLGFYVTLVVTRWWNQYENLPWPDRLMSLVSGFVEGKDEQ 120
EFPLLAVPGPCGRPGSRQARGGSRGAAARGWGGGGGT
GRLLRRTLIRYANLGNVLILRSVSTAVYKRFPSAQHLVQAGFMTPAEHKQLEKLSLPHNM 180
PAAGRRLSVGKGADCSQRN* 139
FWVPWVWFANLSMKAWLGGRIRDPILLQSLLNEMNTLRTQCGHLYAYDWISIPLVYTQVV 240
TVAVYSFFLTCLVGRQFLNPAKAYPGHELDLVVPVFTFLQFFFYVGWLKVAEQLINPFGE 300
DDDDFETNWIVDRNLQVSLLAVDEMHQDLPRMEPDMYWNKPEPQPPYTAASAQFRRASFM 360
GSTFNISLNKEEMEFQPNQEDEEDAHAGIIGRFLGLQSHDHHPPRANSRTKLLWPKRESL 420
LHEGLPKNHKAAKQNVRGQEDNKAWKLKAVDAFKSAPLYQRPGYYSAPQTPLSPTPMFFP 480
LEPSAPSKLHSVTGIDTKDKSLKTVSSGAKKSFELLSESDGALMEHPEVSQVRRKTVEFN 540
LTDMPEIPENHLKEPLEQSPTNIHTTLKDHMDPYWALENRSVLHLNQGHCIALCPTPASL 600
DEAHS* 584
ALSLPFLHNFLGFHHCQSTLDLRPALAWGIYLATFTGILGKCSGPFLTSPWYHPEDFLGP 660
GEGR* 664
|
D