![]() Figure 2 of
Sullivan, Mol Vis 2002;
8:102-113.
Figure 2 of
Sullivan, Mol Vis 2002;
8:102-113.
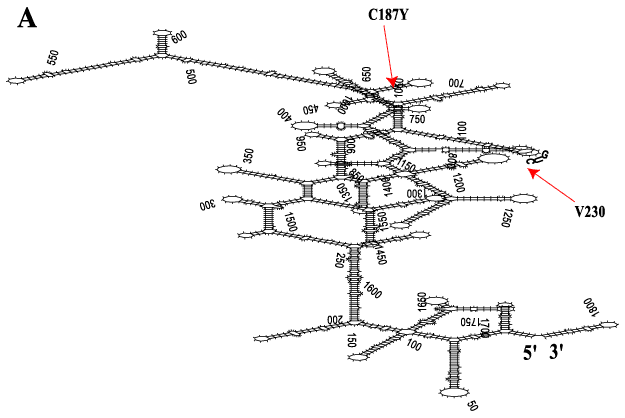
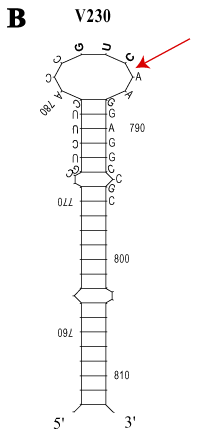
Figure 2. Most stable full length human rod opsin mRNA secondary structure
Full length human rod opsin mRNA from nt 1 (start of transcription) and extending 311 nt beyond the first polyadenylation signal (total 1821 nt length) [46] was subjected to secondary structure folding (RNAFOLD, GCG algorithm) and displayed with SQUIGGLES (GCG). A: The most stable secondary structure with the lowest free energy (DG=-587.9 kCal/mol) is shown without attempting to rotate regions of overlap. Over 90% of the mRNA is hybridized. Part of the predicted stable stem-loop structure that embraces the V230 codon and the target-annealing region of anti-V230 KD hRzs is annotated by sequence. The cleavage target (GUC`) is in bold and the arrow (red) shows the location of the phosphodiester bond broken by anti-V230 hRzs. The adRP C187Y mutation (site not targeted) is shown for reference and is buried in a stem structure. B: Magnification of the stem-loop embracing the V230 codon (GUC`) shows the sequence that hybridizes to the AS flanks of the first/second generation KD hRzs.