![]() Figure 3 of
Hartman, Mol Vis 2001;
7:95-100.
Figure 3 of
Hartman, Mol Vis 2001;
7:95-100.
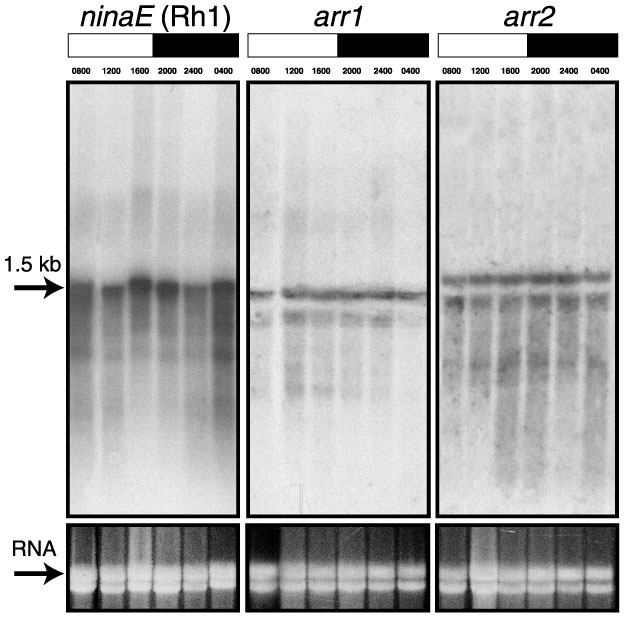
Figure 3. Northern blot analysis
Northern blot analysis reveals expected transcripts of 1.5 kb for ninaE (Rh1) [41,62], 1.45 kb for arr1 and 1.65 kb for arr2 [42,63,64]. The flies were maintained on a 12L/12D cycle. Heads were collected at 4 h intervals: 0800, 1200, 1600, 2000, 2400, and 0400. The lights were on at 0800 and off at 2000. White boxes represent the time of lights on and black boxes represent lights-off. 10 mg of total RNA was loaded per lane and 18S rRNA was used as a loading control. Each band was scanned three times and the peak area was integrated with Sigmagel software. The mean areas were normalized by calculating the ratio of the mean area and the amount of rRNA loaded.
Normalized areas by time (mean±standard deviations)
Gene 0800 1200 1600 2000 2400 0400
----- ------------ ---------- ---------- ---------- ---------- ----------
ninaE 14,991± 312 15,327±815 15,392±452 17,561±317 14,988±818 16,282±538
arr1 9,737±1,066 10,617±257 9,897±371 11,622±188 12,221±918 12,815±247
arr2 12,111± 407 11,666±275 13,948±303 15,065±113 15,365±483 13,383± 11
|