![]() Figure 2 of
Gross, Mol Vis 2000;
6:40-50.
Figure 2 of
Gross, Mol Vis 2000;
6:40-50.
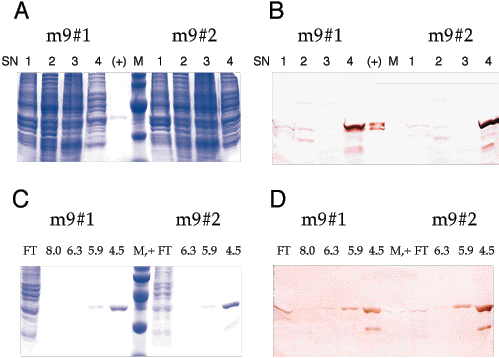
Figure 2. Typical mutant 9 purification
Two separately grown preparations, Mutant 9 number 1 and Mutant 9 number 2, showing supernatants 1 to 4 of the protein preparations and the results of nickel ion affinity purification of SN4 from each preparation. Mutant 9 number 1 and number 2 are individual protein preparations of separate clones with the Mutant 9 G239T substitution. Protein preparation supernatants 1 through 4 (SN 1 to 4) were prepared by differential extraction and centrifugation yielding the several supernatants and a final pellet. An immunoblot with monoclonal antibody H3B5 found Mutant 9 protein mostly in SN4 (urea). Nickel ion affinity purification of SN4 from both Mutant 9 clones, and detection with H3B5 located the mutant protein eluting at pH 5.9 and 4.5, but not with the preceding washes of the column at pH's of 8.0 or 6.3. EcR1 and the mutant EcR1 protein bands ran between the ovalbumin 43 kDa and carbonic anhydrase 29 kDa molecular weight standards. SlyD runs anomalously on SDS PAGE gels [25] and here migrated about the same distance as the 18.4 kDa b-lactoglobulin marker band. According to the manufacturer, the three marker bands mentioned above have apparent molecular weights on gels of 44.7, 28.3 and 19.3 kDa, respectively. Contamination with SlyD was especially evident in the Mutant 9 number 2 pH 4.5 elution fraction on both the SDS PAGE gel and the immunoblot. FT means column flow through, (+) represents EcR1 used as a positive control and M is the marker lane. In Panels A and B, 1 through 4 are the protein preparation supernatants, and in panels C and D, the numbers 8.0, 6.3, 5.9 and 4.5 designate the pH of the denaturing solution used to wash the column or elute the protein from the column. Panels A and C are Coomassie Brilliant Blue stained SDS PAGE gels, and B and D are monoclonal antibody H3B5 stained western blots of samples corresponding to the gels in A and C.