![]() Figure 3 of
Ayyagari, Mol Vis 1999;
5:13.
Figure 3 of
Ayyagari, Mol Vis 1999;
5:13.
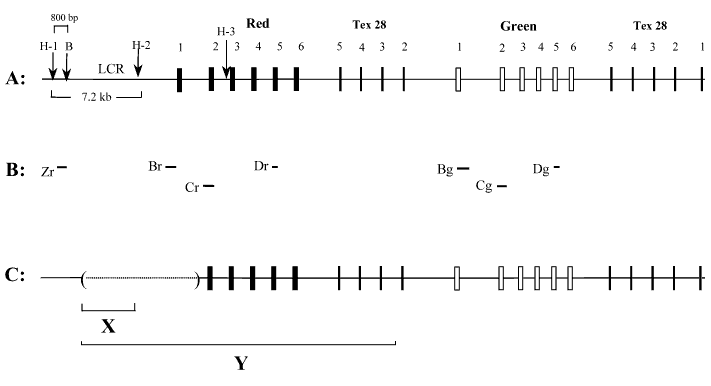
Figure 3. Organization and deletions of the red, green and Tex 28 genes, and probes for Southern blot analysis
A. Organization of the red, green and Tex 28 genes in normal individuals. Boxes indicate the position of exons and numbers above the boxes indicate the number of the exons. Hind III sites in the upstream red gene region and in intron 2 of the red gene are labeled as H-1, H-2, and H-3. "B" represents a Bam HI site. B. Position of the probes used for Southern blot analysis. Zr is a 800 bp probe corresponding to the sequence located between the Hind III and Bam HI sites in the upstream red gene region. Br and Bg hybridize to exon 1 of the red and green genes respectively. Cr and Cg correspond to exon 2 of the red and green genes and Dr and Dg represent exon 5 of the red and green genes respectively. C. Structure of the red, green and Tex 28 genes in this family with macular atrophy. Solid line indicates the region that is present. The dotted line in brackets indicates the deleted region. The position of the sequence shown in C corresponds to the structure of red, green and Tex 28 genes shown in A. X and Y represent the position of the deletion in families E:HS106 [10] and HS129 [11], respectively, that are reported to have maculopathy.