![]() Figure 7 of
Rozsa, Mol Vis 4:20, 1998.
Figure 7 of
Rozsa, Mol Vis 4:20, 1998.
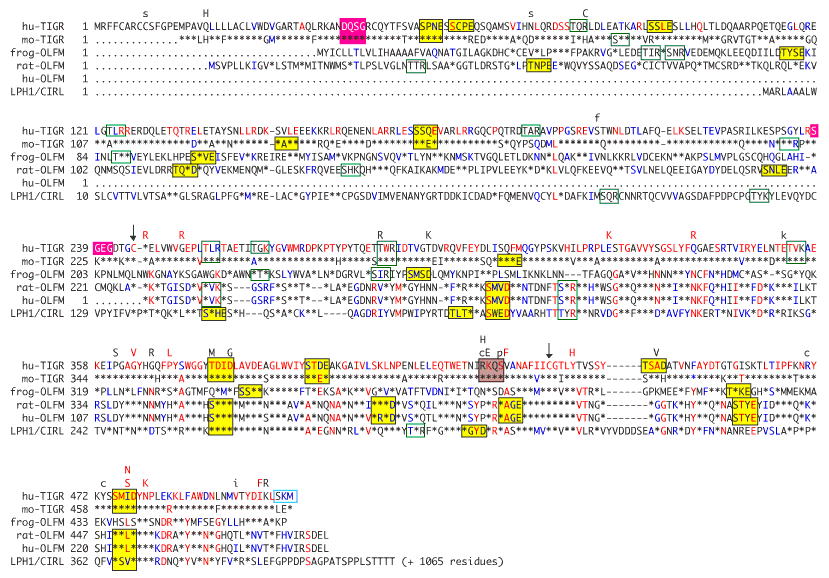
Figure 7. Amino acid alignment of olfactomedin-like proteins
Human TIGR/MYOC (hu-TIGR, U85257) [25], mouse TIGR/MYOC (mo-TIGR, AF049791, AF049792, AF049793, AF049794, AF049795, AF049796) [23,43], frog olfactomedin (frog-OLFM) (L13595) [45], rat olfactomedin-like (rat-OLFM) (U03417) [46], human olfactomedin-like (hu-OLFM) (U79299) [47], and rat latrophilin (LPH1/CIRL) (U78105) [41] proteins were compared using the PILEUP program. Amino acids are indicated by single-letter code and are numbered according to their ungapped sequence. Within the alignment, gaps introduced to optimize alignment are shown as dashes, conserved substitutions are shown in blue, highly conserved changes are shown in red, and residues identical to the human TIGR/MYOC sequence are shown by asterisks (*). The location of TIGR/MYOC missense mutations are shown above the alignment in uppercase letters. Mutations associated with juvenile-onset glaucoma are shown in red, uppercase letters. Mutations considered unlikely to cause disease by Alward et al [26] are in lowercase. Motifs identified from the Prosite database are boxed in green outline, yellow, red, gray and blue for protein kinase C, casein kinase II phosphorylation, glycosaminoglycan initiation attachment, cAMP dependent protein phosphorylation, and peroxisome C-terminal targeting signal motifs, respectively. Conserved cysteines are shown with arrows. Rat latrophilin continues an additional 1065 residues where indicated.