Figure 4. STRING cluster analysis of regulated proteins in DME. A: A large cluster was formed by proteins involved in complement and coagulation cascades, including complement factors (C2,
C3, C4B, C5, C6, C7, C8A, C8G, C9, and CFH), fibrinogen chains (FGA, FGB, and FGG), vitronectin (VTN), alpha-1-antitrypsin
(SERPINA1), antithrombin-III (SERPINC1), heparin cofactor 2 (SERPIND1), alpha-2-antiplasmin (SERPINF1), plasma kallikrein
(KLKB1), kininogen-1 (KNG1), alpha-2-macroglobulin (A2M), and prothrombin (F2). Another cluster was involved in ECM–receptor
interaction, including agrin (AGRN), fibronectin (FN1), dystroglycan (DAG1), and basement membrane-specific heparan sulfate
proteoglycan core protein (HSPG2). A third cluster of proteins was involved in the cholesterol metabolism, including apolipoprotein
A-I (APOA1), apolipoprotein C-III (APOC3), apolipoprotein A-IV (APOA4), low-density lipoprotein receptor-related protein 2
(LRP2), and phosphatidylcholine-sterol acyltransferase (LCAT). B–F: A cluster of proteins represented multiple KEGG pathways. This was the case for fructose-bisphosphate aldolase C (ALDOC),
fructose-bisphosphate aldolase A (ALDOA), alpha-enolase (ENO1), phosphoglycerate kinase 1 (PGK1), L-lactate dehydrogenase
A chain (LDHA), L-lactate dehydrogenase B chain (LDHB), and pyruvate kinase PKM (PKM). The regulation of these proteins suggests
that DME was associated with glycolysis/gluconeogenesis, the pyruvate metabolism, the carbon metabolism, HIF signaling, and
the glucagon signaling pathway.
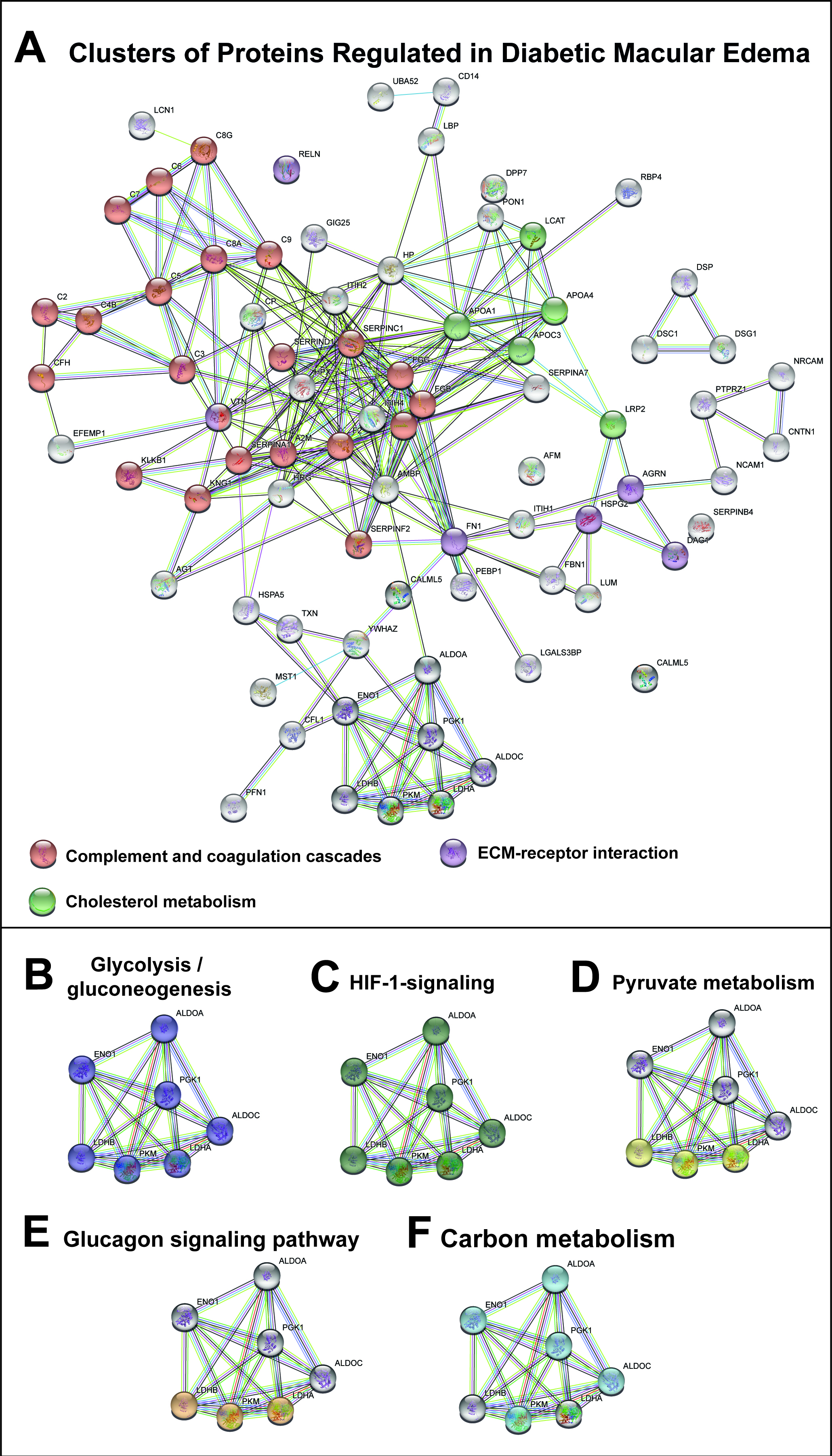
 Figure 4 of
Cehofski, Mol Vis 2024; 30:17-35.
Figure 4 of
Cehofski, Mol Vis 2024; 30:17-35.  Figure 4 of
Cehofski, Mol Vis 2024; 30:17-35.
Figure 4 of
Cehofski, Mol Vis 2024; 30:17-35. 