Figure 1. Presentation of DEPs. A: Number of DEPs. B: Hierarchical cluster analysis of the DEPs (Columns represent samples, rows represent proteins, and the shorter the distance
of cluster branches, the higher the similarity of proteins). Red represents upregulated proteins, blue represents downregulated
proteins, and gray represents no protein quantitative information. C: DEPs volcano plot showed that these were distributed in two different quadrants (the logarithm of the fold change of each
quantifiable protein was taken as base 2, and the negative logarithm of the p value was taken as base 10). The horizontal
axis represents the fold change (log2 value) of the differential protein, and the vertical axis represents the p value (-log10
value). Gray dots represent proteins with insignificant differences, red dots represent upregulated proteins, and blue dots
represent downregulated proteins. The top 10 protein was selected to display. D: Box plot of 9 DEPs. *** p<0.001, ** 0.001<p<0.01, * 0.01<p<0.05.
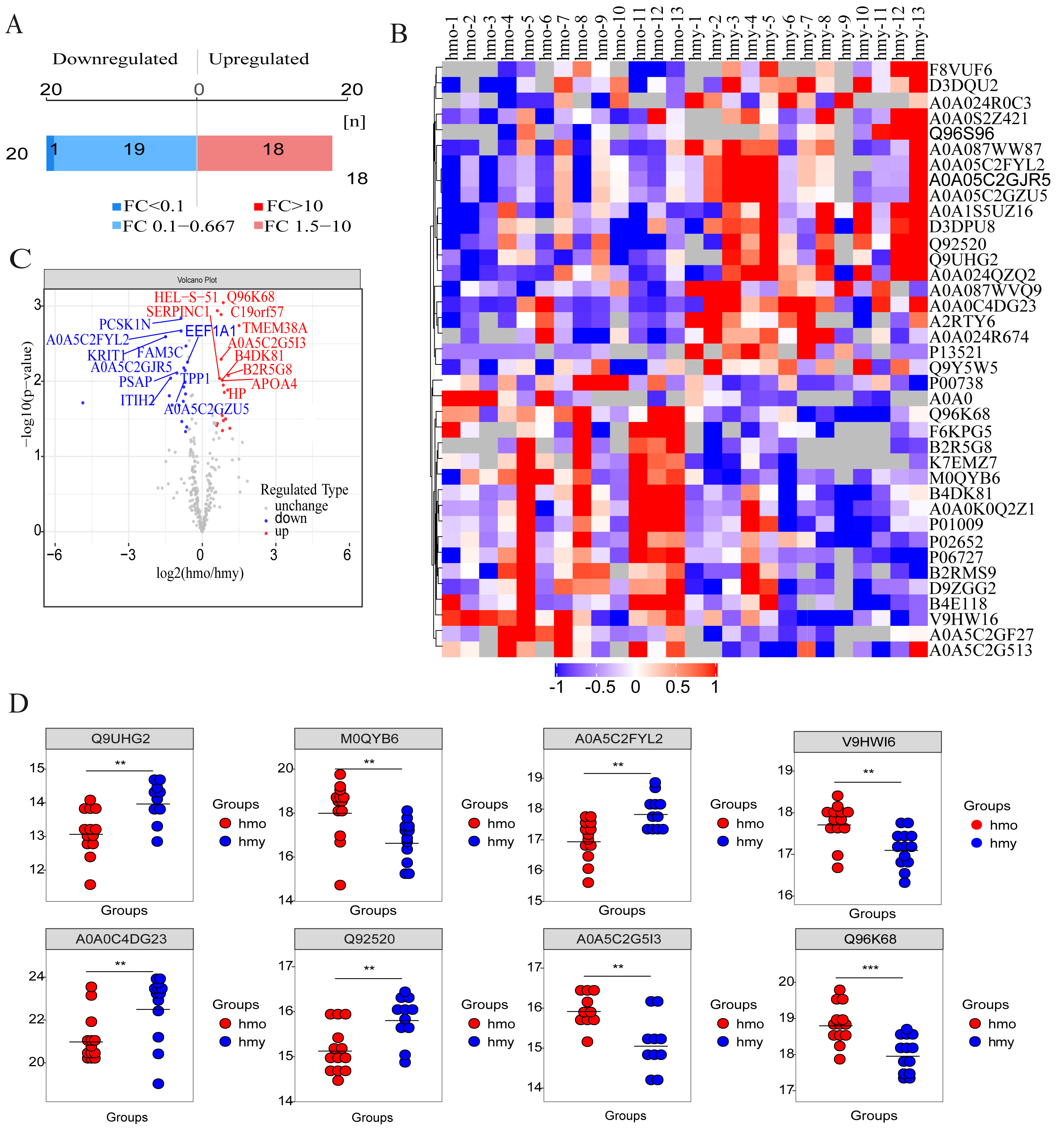
 Figure 1 of
Wen, Mol Vis 2024; 30:137-149.
Figure 1 of
Wen, Mol Vis 2024; 30:137-149.  Figure 1 of
Wen, Mol Vis 2024; 30:137-149.
Figure 1 of
Wen, Mol Vis 2024; 30:137-149. 