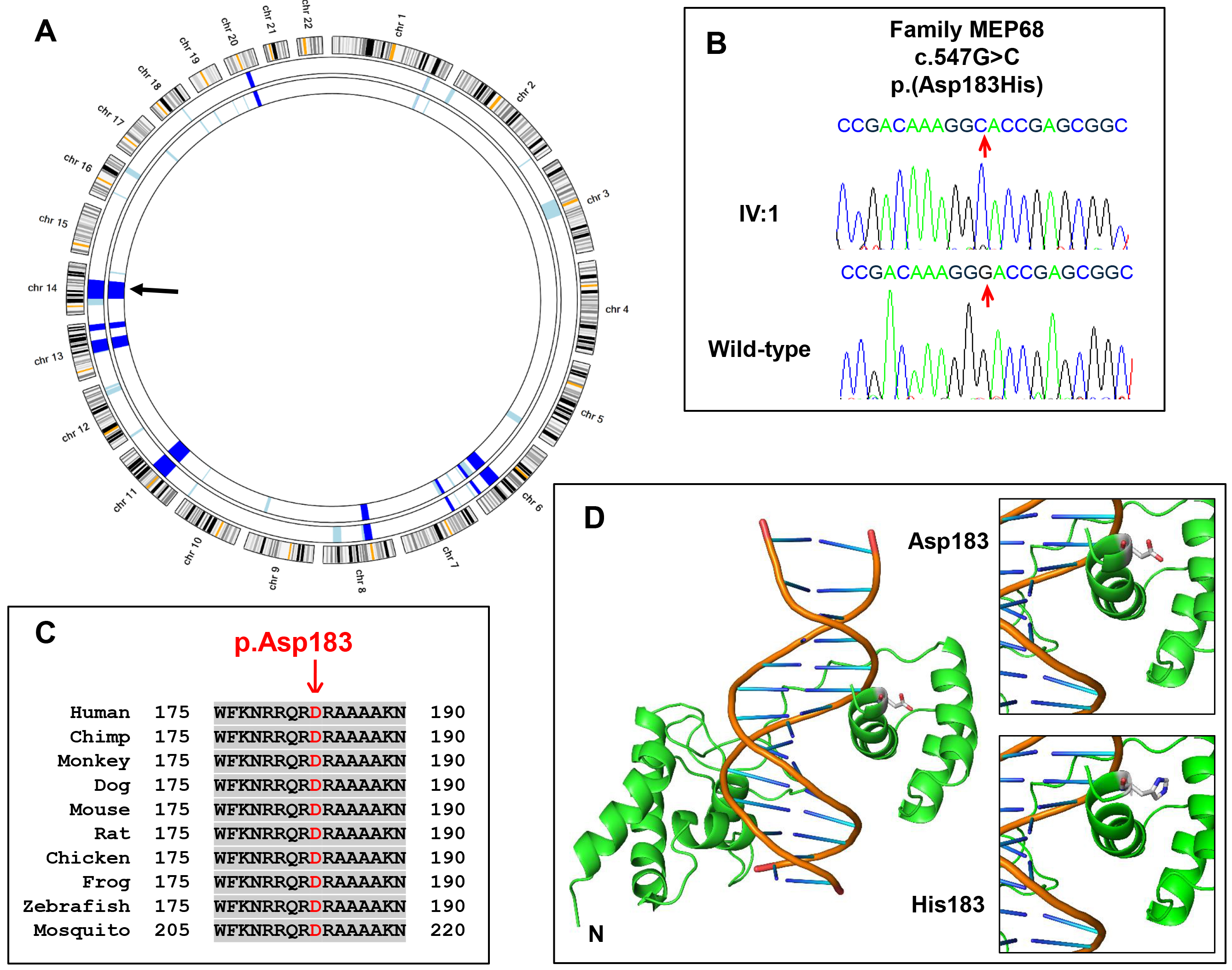
Figure 3. Genetic analysis of family MEP68.
A: AgileMultiIdeogram representation of the homozygous regions from SNP-genotyping data of two affected individuals (IV:1 &
IV:3). Homozygous regions are highlighted in light blue, and common autozygous regions are highlighted in dark blue.
SIX6 (arrow) maps to a common 42 Mb homozygous region on chromosome 14 (33,856,948–75,827,741, GRCh37, hg19).
B: Sequence electropherograms highlighting the nucleotide substitution mutation, c.547G>C, in an affected individual from the
family and a normal control subject.
C: Multiple protein sequence alignment of human SIX6 with orthologs surrounding the p.D183 residue. Accession numbers: human,
NP_031400.2; chimp,
XP_522870.3; monkey,
XP_014999313.1; dog,
XP_547840.3; mouse,
NP_035514.1; rat,
NP_001101502.1; chicken,
NP_001376294; frog,
NP_001093696.1; zebrafish
NP_957399.1; mosquito,
XP_003436640.1.
D: Modeling of SIX6 wild-type and p.(Asp183His) mutant. Ribbon representation of the structural model of SIX6 protein (green)
bound to DNA (orange). The N-terminus and the atom types at the 183
rd residue (white=carbon; blue=nitrogen; red=oxygen) are highlighted. Note the location of the 183
rd residue sitting in the major groove of double-stranded DNA. It is likely that the change from a negatively charged D183 amino
acid in the wild-type sequence to a positively charged H183 in the mutant is likely to disrupt this protein-DNA interaction.
 Figure 3 of
Panagiotou, Mol Vis 2022; 28:57-69.
Figure 3 of
Panagiotou, Mol Vis 2022; 28:57-69.  Figure 3 of
Panagiotou, Mol Vis 2022; 28:57-69.
Figure 3 of
Panagiotou, Mol Vis 2022; 28:57-69.