Figure 1. Design of molecular inversion probe and MIP-based target-sequencing workflow. A: Molecular inversion probe (MIP) structure and design for regions of interest. The general structure for the MIP is a common
41-bp linker flanked by a target-specific extension and a ligation arm of 16 to 28 bp. B: MIP-based target-sequencing workflow. (i) Genomic DNA was sheared into 300–500 bp fragments. (ii) Pooled probes are added
to the genomic DNA, and each probe is hybridized to the target DNA sequences through the target-specific sequences on both
ends. (iii) The gaps are filled and circularized with DNA polymerase and ligase. (iv) The linear genomic DNA and uncircularized
padlock probes are removed with exonuclease. (v) The circularized probes are amplified with a universal sequencing primer
and a sample barcode primer.
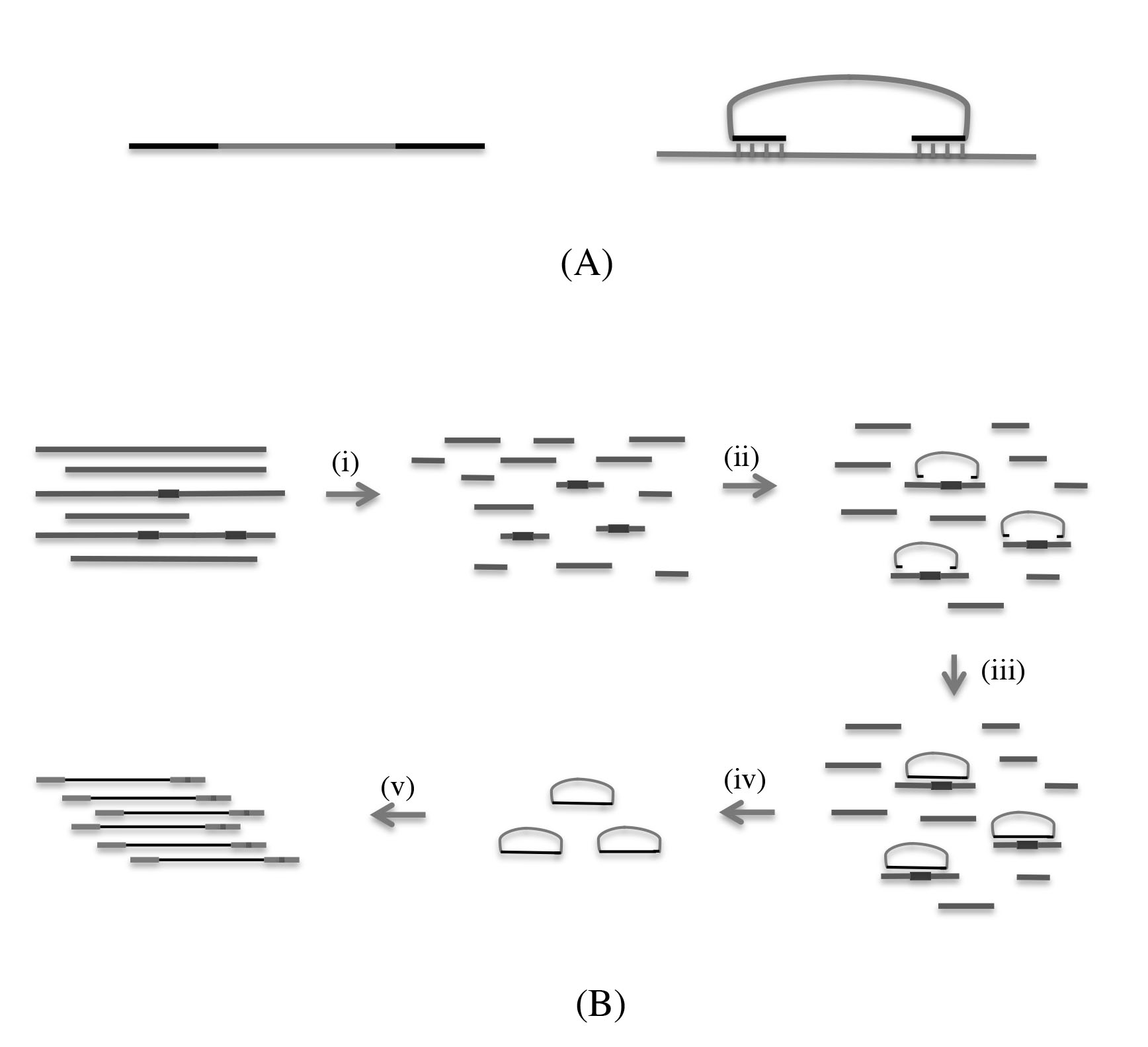
 Figure 1 of
Liu, Mol Vis 2020; 26:378-391.
Figure 1 of
Liu, Mol Vis 2020; 26:378-391.  Figure 1 of
Liu, Mol Vis 2020; 26:378-391.
Figure 1 of
Liu, Mol Vis 2020; 26:378-391. 