Figure 7. In silico pathogenicity predictions for the novel GSN (c.1631T>G; p.Met544Arg) variant identified in this study. A: Variant impact was assessed using the Sorting Intolerant From Tolerant (SIFT), Polymorphism Phenotyping v2 (PolyPhen-2),
MutationTaster, and Protein Variation Effect Analyzer (PROVEAN) algorithms. B: In silico prediction of the effects on the protein stability produced by the p.Met544Arg replacement on GSN. Algorithm predictions
are based on changes in folding free energy (ΔΔG in Kcal/mol). Protein stability predictions were performed using the crystal
structure of calcium-free human gelsolin. C: Alignment of the amino acid sequence of gelsolin across different species. The shaded column with the red mark indicates
the position of the mutated residue identified in this study. As observed, methionine 544 is strictly conserved among numerous
vertebrate species.
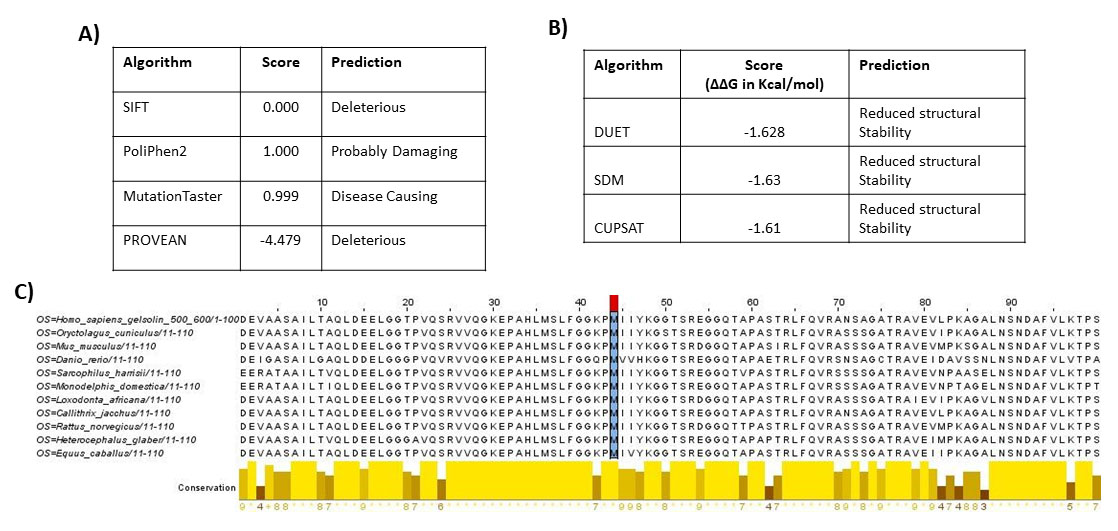
 Figure 7 of
Cabral-Macias, Mol Vis 2020; 26:345-354.
Figure 7 of
Cabral-Macias, Mol Vis 2020; 26:345-354.  Figure 7 of
Cabral-Macias, Mol Vis 2020; 26:345-354.
Figure 7 of
Cabral-Macias, Mol Vis 2020; 26:345-354. 