Figure 4. Identified genetic variants. A: Domains of the CFH protein that is composed of 20 repetitive units of 60 amino acids called short consensus repeats (SCRs)
or complement control protein modules (CCPs). The amino acid sequence of CCPs 12 (blue color) and 13 (red color) are depicted
(690–804). The eight residues that are in the black box are located in the CCP 12 and 13 linker region. The pink colored box
shows amino acid residues that were found to be deleted in our study. B: Chromatograms of a homozygous wild type (top) and a heterozygous affected individual (bottom) with the c.2247_2252del; p.(Leu750_Lys751del)
CFH variant. Arrows represent the beginning and the end of the six deletion nucleotides. C: Illustration of reported variants in chromosome 6: chr6:100020205–100143306 duplication, 100,040,906 G>T, 100,040,987 G>C,
and 100,041,040 C>T. The red arrow represents the location of the variant identified in the current study within the previously
reported hotspot region- a DNase hypersensitivity site (blue), upstream of PRDM13 gene (green). Genomic localization is based on human reference sequence hg19 (GRCH37). D: Chromatograms of a homozygous wild-type (top) and a heterozygous affected individual (bottom) with the chr6: 100040974A>C
variant.
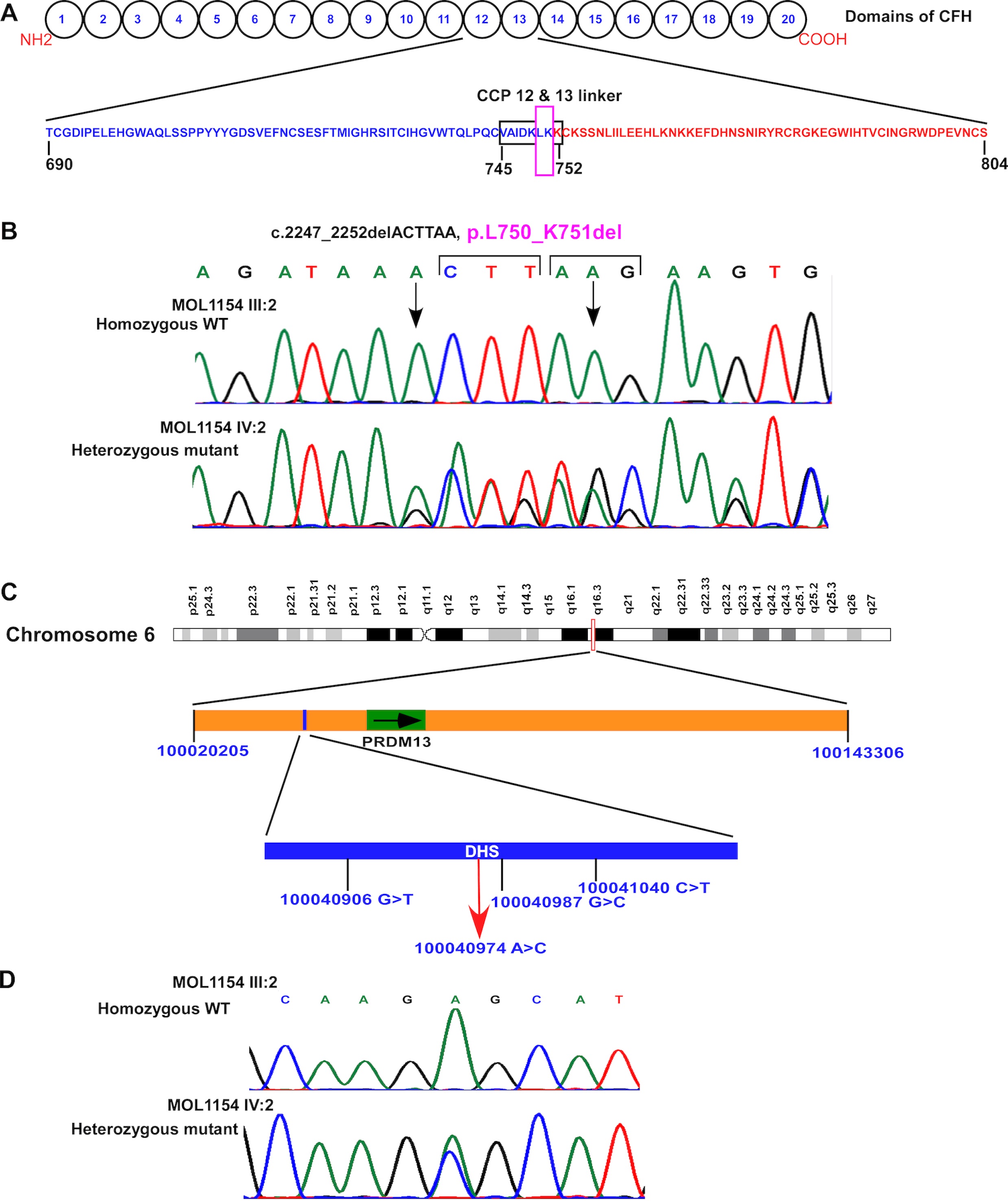
 Figure 4 of
Namburi, Mol Vis 2020; 26:299-310.
Figure 4 of
Namburi, Mol Vis 2020; 26:299-310.  Figure 4 of
Namburi, Mol Vis 2020; 26:299-310.
Figure 4 of
Namburi, Mol Vis 2020; 26:299-310. 