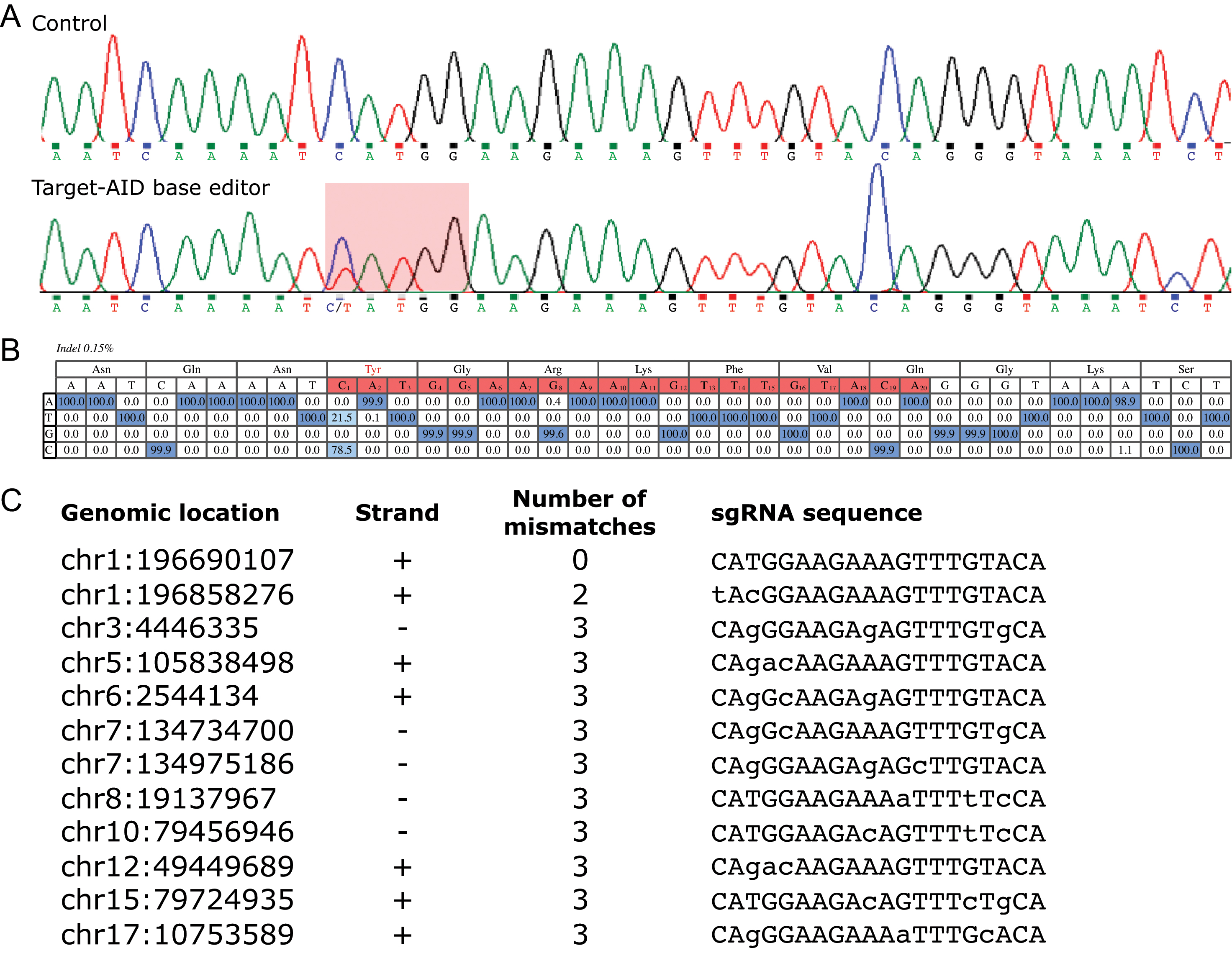
Figure 2. Editing efficiency of the Target-AID base editor at Y402H
CFH variant (
rs1061170).
A: Sanger sequencing chromatograms showing the target locus. Control refers to HEK293A-CFH(p.His402) cells that have been treated
with a combination of the Target-AID base editor with a sgRNA targeting
LacZ. The second chromatogram displays results following transfection with the Target-AID base editor and a sgRNA specific to
the
rs1061170 region. The highlighted region is the putative activity window of Target-AID showing the incidence of a C:G to T:A nucleotide
correction.
B: Base calling for targeted next-generation deep sequencing of the base-editor treated HEK293A-CFH(p.His402) cells. Percentages
represent the fraction of sequencing reads occupied by a particular nucleotide at the specified position. The target protospacer
is highlighted in red and each nucleotide is numbered 1 to 20 in which the PAM consensus sequence is considered positions
+21 to +23. Deep sequencing shows that 21.5% of the target cytosines were edited into thymine nucleotides at position C1.
Additionally, the percentage of indel formation is 0.15% (amplicon results of three independent biologic replicates).
C: List of sgRNA sequences screened at the top 11 off-target sites that also contain at least one cytosine in the putative
Target-AID activity window in the presence of a NGG PAM site. Each off-target site was evaluated using next generation deep
sequencing and no off-target base editing was identified.
 Figure 2 of
Tran, Mol Vis 2019; 25:174-182.
Figure 2 of
Tran, Mol Vis 2019; 25:174-182.  Figure 2 of
Tran, Mol Vis 2019; 25:174-182.
Figure 2 of
Tran, Mol Vis 2019; 25:174-182.