Figure 1. Activity of different base editors targeting the rs1061170 SNP. The base editor variant, sgRNA protospacer and PAM consensus
nucleotide, and incidence of base editing are indicated. Base editing was determined if a peak was observed at the risk variant
allele following analysis of Sanger sequencing chromatograms compared to the LacZ negative control (n = 3 biologic replicates performed on different days for all base editor constructs). PAM sequences and
gRNA target sequences are shown in orange and blue, respectively. Putative base editor activity windows are displayed in green,
where dark green indicates higher editing efficiency and light green denotes lower editing efficiency.
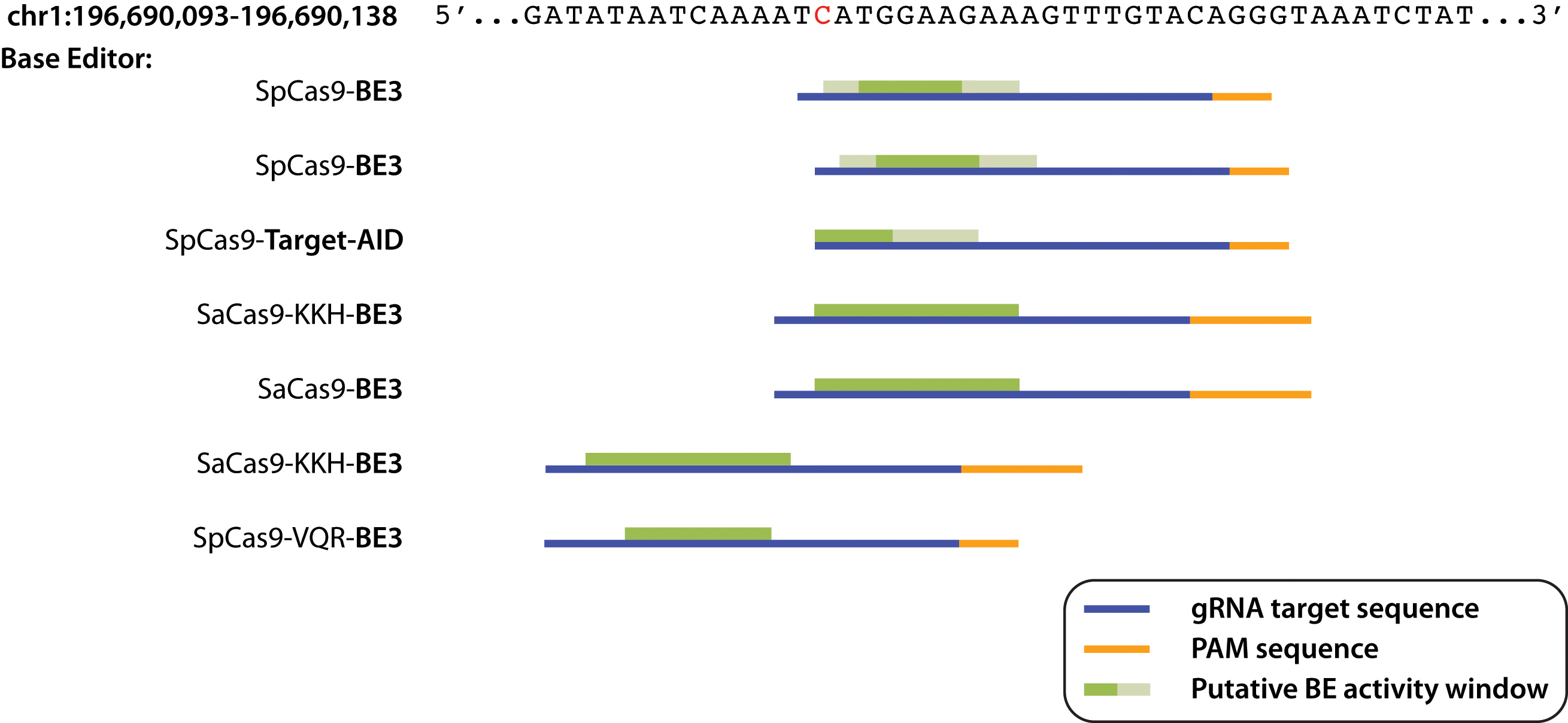
 Figure 1 of
Tran, Mol Vis 2019; 25:174-182.
Figure 1 of
Tran, Mol Vis 2019; 25:174-182.  Figure 1 of
Tran, Mol Vis 2019; 25:174-182.
Figure 1 of
Tran, Mol Vis 2019; 25:174-182. 