Figure 1. The Rax C-terminal region contains activation and repression domains. Full-length (FL) or mutated Rax or Rax2 was assayed
for the ability to activate expression of a Xenopus rhodopsin gene promoter–luciferase reporter gene (C and D or E, respectively). A: Schematic representation of the Rax constructs assayed. Numbers indicate amino acid residues. Dotted lines indicate internal
deletions. Point mutations are marked by an asterisk (*). B: Schematic representation of Rax2 constructs assayed. C, D: Luciferase assay results for Rax constructs diagrammed in A. Reporter gene activity is calculated as firefly luciferase activity normalized to the Renilla luciferase control and is presented relative to activity of a pCS2-only plasmid control. Values represent the mean of three
to nine independent experiments (nine replicates per experiment). Error bars represent standard deviation from the mean. All
vector-only and Rax activity values are statistically significantly different from full-length Rax activity using a corrected
(Bonferroni) critical value of p<0.0017 (*p=4.06 × 10−48, n=81; **p=3.25 × 10−8, n=27; ***p=1.04 × 10−23, n=27; ****p=6.15 × 10−56, n=27; *****p=3.73 × 10−6, n=27; #p=8.75 × 10−18, n=54; ##p=2.42 × 10−10, n=27; ###p=3.28 × 10−20, n=27). E: Rax2 ΔC180 activity is not statistically significantly different from full-length Rax2 activity (Student t test, p=0.02). Abbreviations: HD = homeodomain, OAR = orthopedia, aristaless, rax, OP = octapeptide domain, PPXY = PPXY motif
(putative YAP/TAZ binding sequence), RX = Rx domain. *in construct 3PA denotes mutated proline residues (to alanine residues).
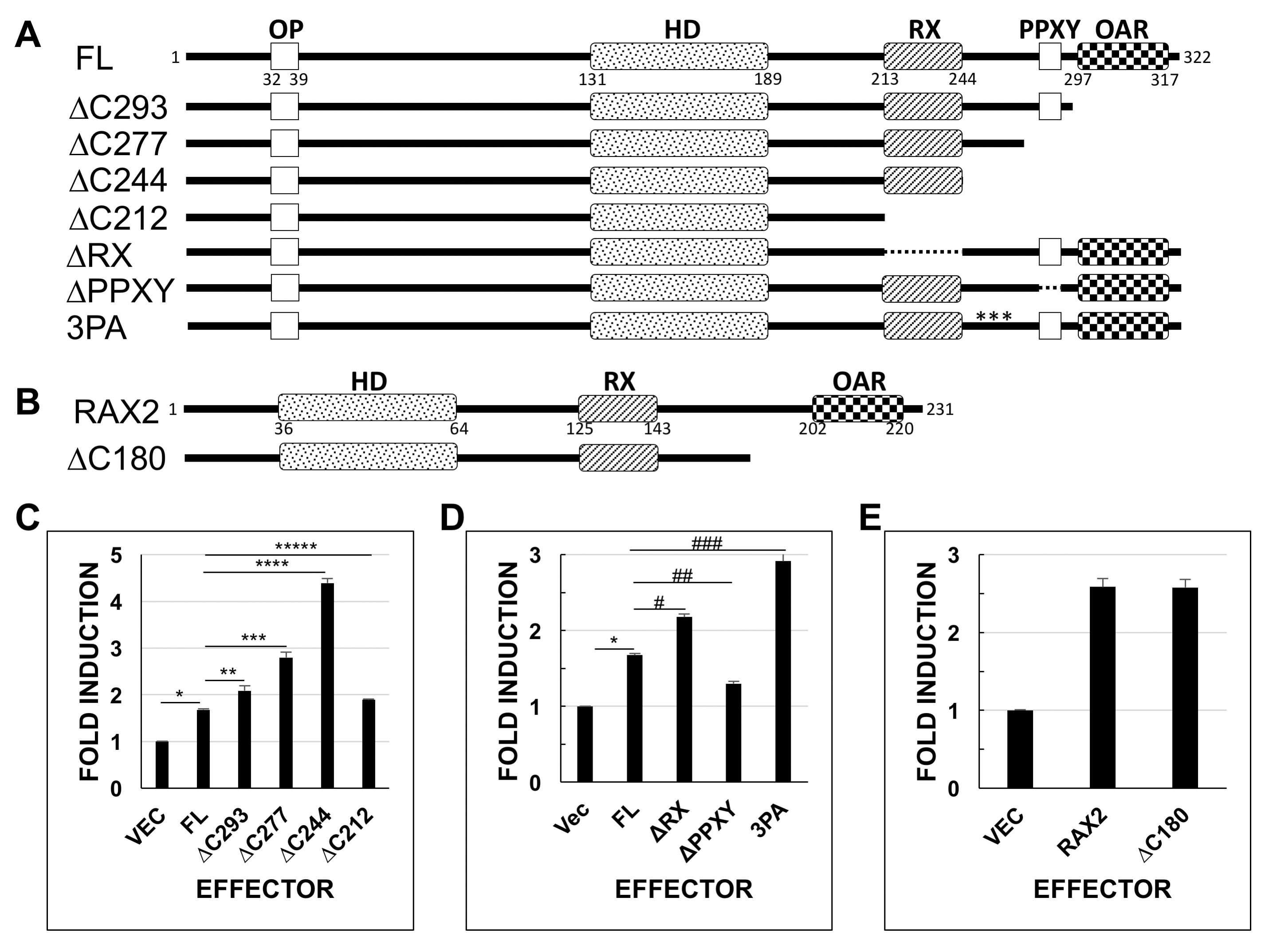
 Figure 1 of
Buescher, Mol Vis 2019; 25:165-173.
Figure 1 of
Buescher, Mol Vis 2019; 25:165-173.  Figure 1 of
Buescher, Mol Vis 2019; 25:165-173.
Figure 1 of
Buescher, Mol Vis 2019; 25:165-173. 