Figure 1. Comparison of the protein and gene structure of vertebrate Dscam and Sdk genes. A: The combination of domain structures for zebrafish Dscama, Dscamb, Dscaml1, Sdk1a, Sdk1b, Sdk2a, and Sdk2b proteins as predicted
from the zebrafish genome. The corresponding structures for Dscam, Sdk1, and Sdk2 for other non-teleost vertebrates are also
displayed. B: Positions of dscam and sdk genes in the zebrafish genome. Chromosome numbers are shown as 1, 3, 10, 12, and 15. Paralogs are highlighted with the same
colors. Scale bar=10 Mbp. C–E: The predicted exon–intron structures for zebrafish dscam and sdk genes obtained from Ensembl (release 90). Rectangles indicate exon locations targeted by RNA antisense probes. Scale bars=20
Kbp. Abbreviations: Dscam=Down syndrome cell adhesion molecule; Sdk=sidekick; Kbp=Kilo base pairs; Mbp=Mega base pairs.
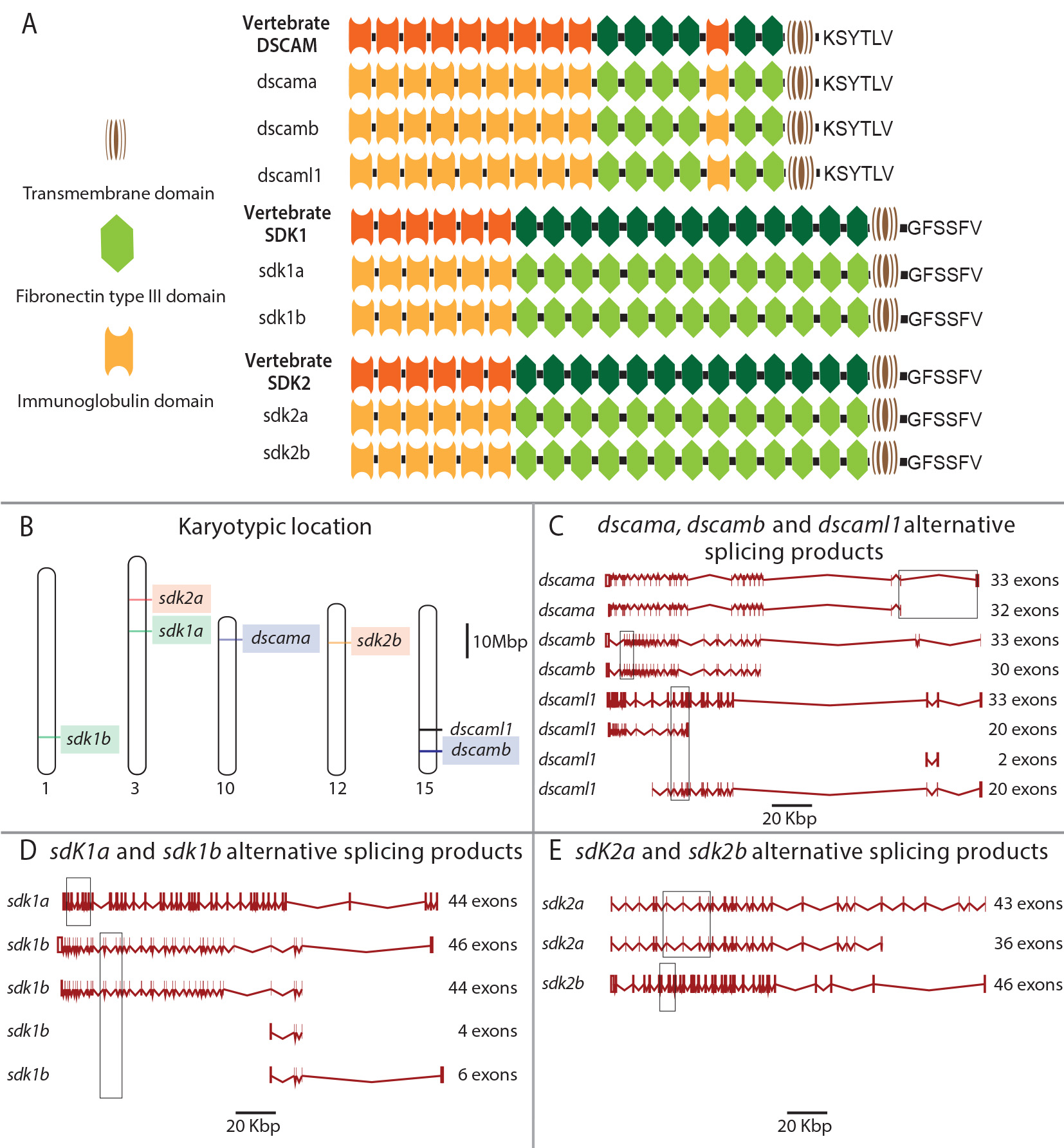
 Figure 1 of
Galicia, Mol Vis 2018; 24:443-458.
Figure 1 of
Galicia, Mol Vis 2018; 24:443-458.  Figure 1 of
Galicia, Mol Vis 2018; 24:443-458.
Figure 1 of
Galicia, Mol Vis 2018; 24:443-458. 