Figure 1. Identifying Orthologous DE Genes Using the Rosetta Stone Ortholog Table. A: A list of DE genes from two rat and one zebrafish data sets being compared. The list has been shortened to facilitate this
example of how the orthologs are identified by the common human ortholog. B: The Rosetta Stone Ortholog Table lists all of the orthologs of each human gene for each species in the table. It is important
to note that an ortholog from a species may appear multiple times in the Rosetta Stone. If a species has no reported human
ortholog, then that index in the table appears as “no orthologue.” C: The lists of DE genes from each data set shown in (A) are now translated into the corresponding human ortholog using an algorithm written in Python and then aligned to show common
DE genes in each data set. This translation allows a direct comparison between data sets between species. In this table, nd
refers genes that were not detected as DE genes in the data set.
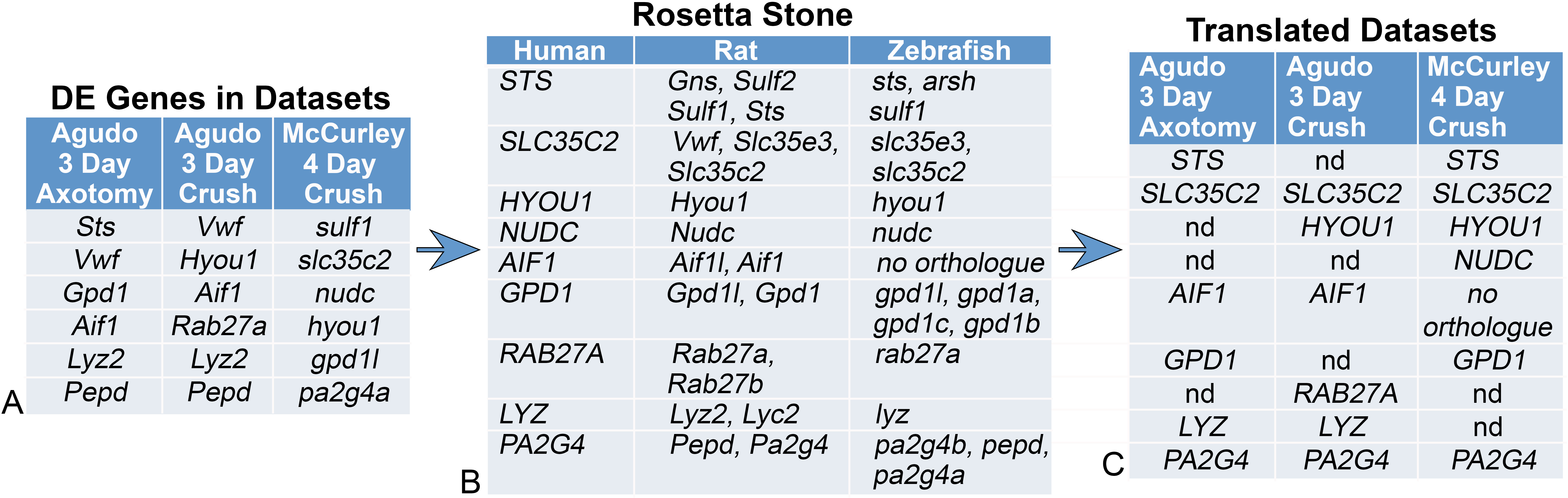
 Figure 1 of
Donahue, Mol Vis 2017; 23:987-1005.
Figure 1 of
Donahue, Mol Vis 2017; 23:987-1005.  Figure 1 of
Donahue, Mol Vis 2017; 23:987-1005.
Figure 1 of
Donahue, Mol Vis 2017; 23:987-1005. 