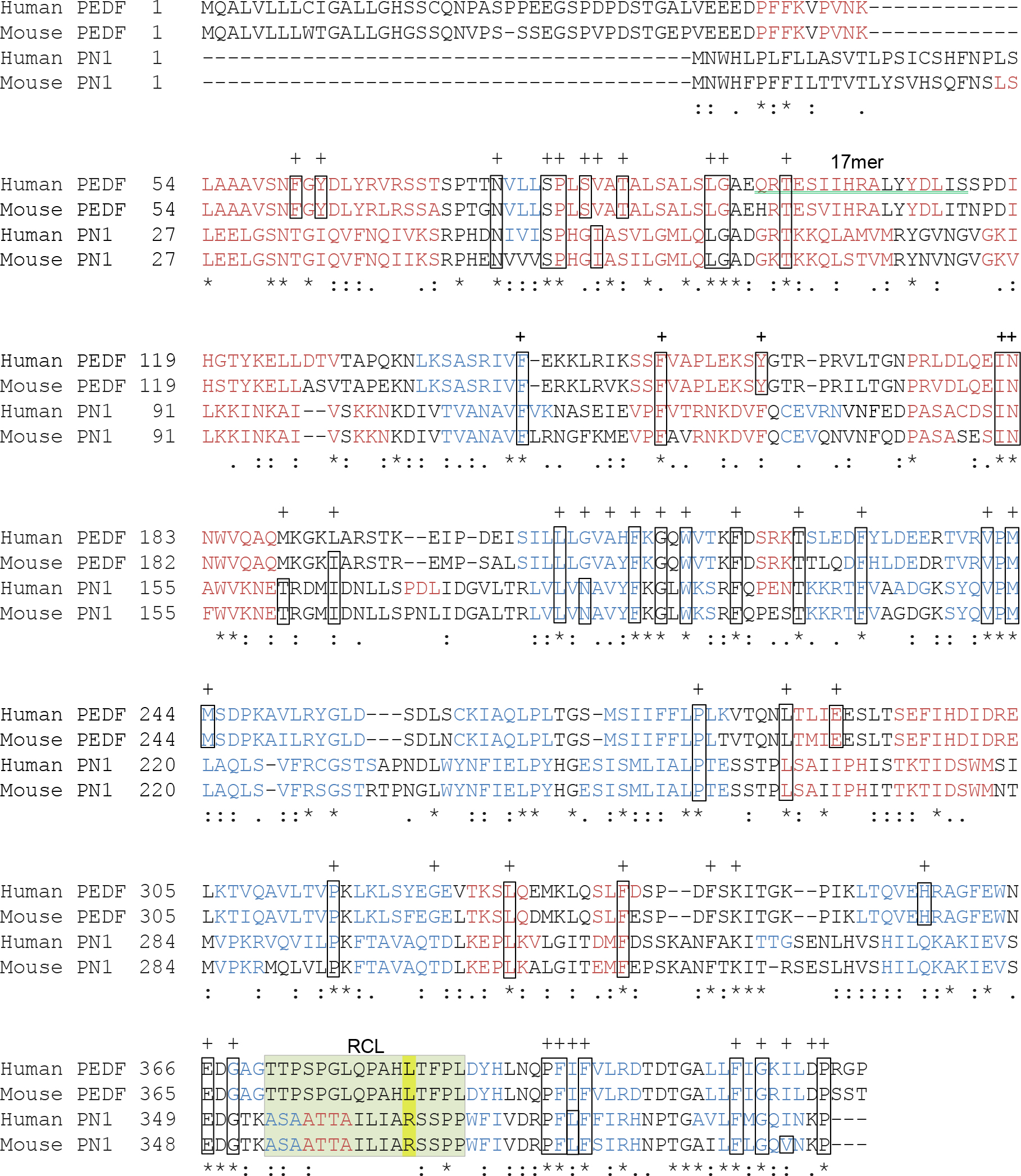
Figure 1. Primary sequence comparison of PN-1 and PEDF. The alignment performed using the
ClustalW2 Multiple Sequence Alignment EMBL-EBI program illustrates the degree to which the amino acid sequences of human and mouse PEDF (
NP_002606.3 and
NP_035470.3, respectively) and PN-1 (
NP_006207.1 and
NP_033281, respectively) are conserved. The secondary structures predicted based on the amino acid sequences are depicted in red (alpha
helices) and blue (beta strands). The sequence of the 17mer fragment of human PEDF that binds to PEDF-R and displays neurotrophic
activity in the retina is underlined with a green double line. Within the reactive center loop (RCL, highlighted in pale green),
P1, the residue that confers protease specificity, is highlighted in yellow. Residue identity (*), residues with strongly
similar properties scoring ˃0.5 in the Gonnet PAM 250 matrix (:) and residues with weakly similar properties scoring ˂0.5
on the Gonnet PAM 250 matrix (.) are indicated. The 51 conserved residues among serpins defined by Huber and Carrell [
28] as key to maintaining the integrity of the serpin spatial structure are indicated with a “+,” and the boxes correspond to
the amino acid found in the archetype serpin anti-trypsin.
 Figure 1 of
Winokur, Mol Vis 2017; 23:372-384.
Figure 1 of
Winokur, Mol Vis 2017; 23:372-384.  Figure 1 of
Winokur, Mol Vis 2017; 23:372-384.
Figure 1 of
Winokur, Mol Vis 2017; 23:372-384.