Figure 5. Developmental time course of Pou4f2Cre activity within the retina. A developmental time course of Cre targeting within the retina was performed by immunostaining
the retinal cryosections with neural markers (PAX6, POU4F2, ChAT, and Calsenilin). The proportions of reporter-positive cells
for each of the markers at P0, P5, P10, P20, and P40 were quantified. Progression in the proportion of cells targeted by Pou4f2Cre increased in all of the markers over developmental time, except the POU4F2-positive cells, which were 100% targeted starting
from P0. A–D: Developmental time course of PAX6 except P20. E–H: Representative images of all the markers at P20. I: Graph illustrating the proportion of cells positive for the Ai9 reporter over time. PAX6-positive cells were subdivided
into cells found within the retinal ganglion layer (RGL), the inner nuclear layer (INL), and horizontal cells (HCs) in the
outer INL. Calsenilin (Csen) was used to label type 4 bipolar cells, and bipolar cells were the only quantified cell type
reported for this stain. J–M: Cre protein staining. J: Cre was detected within the cell bodies and neurites of cells in the RGL (arrowheads) and horizontal cells (arrow). K: Cre negative control. The Cre protein was not observed. L: Cre was detected within the nuclei and neurites of cells within the RGL but was absent from most nuclei within the INL at
P10 (L’). M: Cre was detected within the nuclei of cells in the RGL and the INL, consistent with reporter activation. M’: Inset showing Cre within cells of the INL. N: Pou4f2Cre effectively targets a conditional allele of Dscam at P45 in bipolar cells, evidenced by accumulation of the DSCAM protein in the cell body. Abbreviations: Csen = calsenilin.
Scale bar in H = 20 μm. Scale bars in J–L = 50 μm. Scale bar in M = 20 μm. Insets in J–K = 25 μm box.
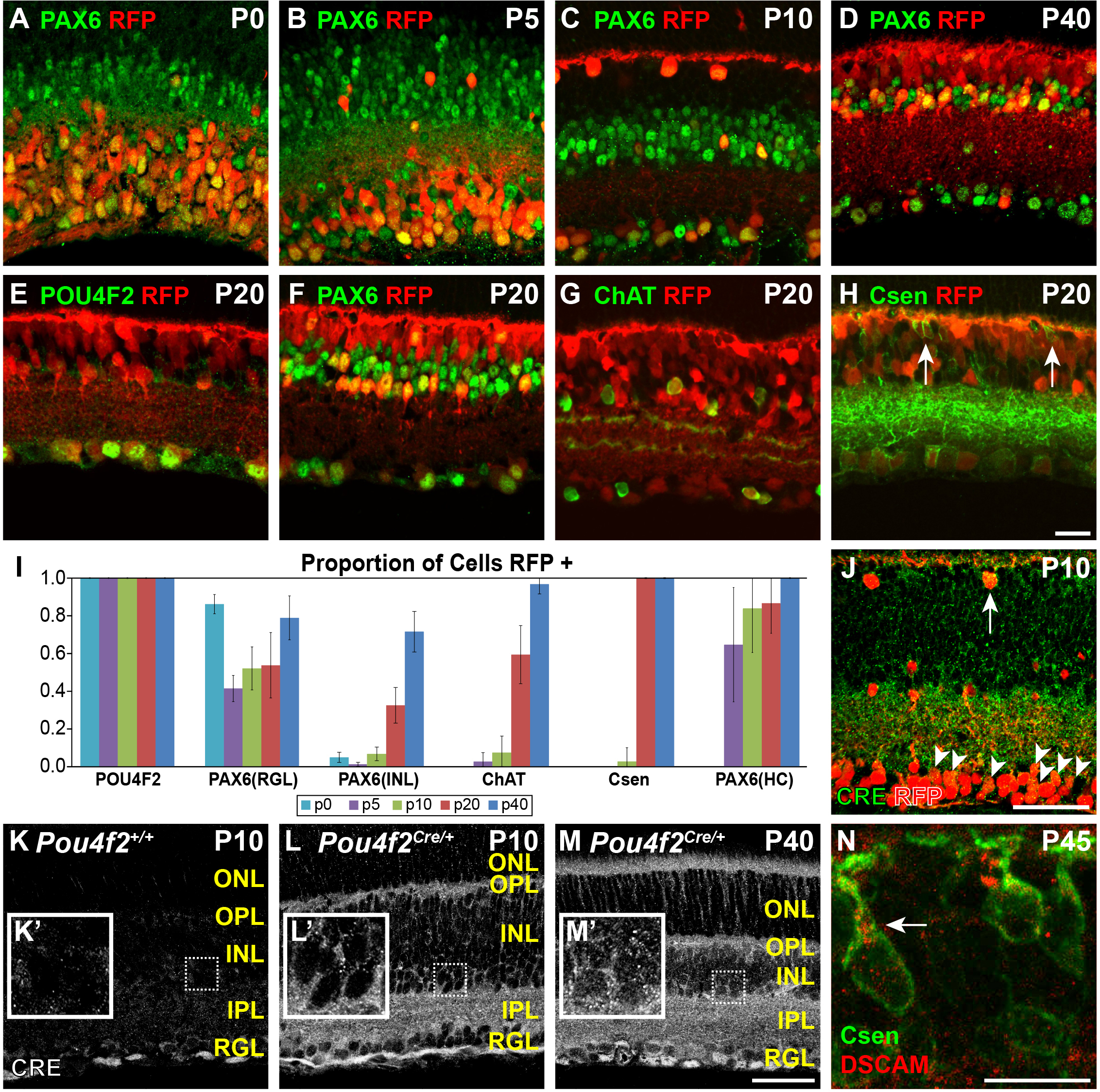
 Figure 5 of
Simmons, Mol Vis 2016; 22:705-717.
Figure 5 of
Simmons, Mol Vis 2016; 22:705-717.  Figure 5 of
Simmons, Mol Vis 2016; 22:705-717.
Figure 5 of
Simmons, Mol Vis 2016; 22:705-717. 