Figure 5. Identification of a splice donor site mutation in RP1. A: Illustration of the deletion breakpoints that remove all three coding exons of RP1.B: Forward sequence chromatograms of bases flanking the homozygous deletion. C: Reverse sequence chromatograms of bases flanking the homozygous deletion. D: In silico analysis of the splice donor site mutation in RP1. The HSF2 algorithm predicted a consensus value (CV) of 80.96 for the wild-type splice donor site (c.787+1G) and 48.29 (c.787+1A)
for the mutant. The consensus value deviation of −32.67 suggests that the loss of the wild-type splice site will result in
retention of intron 3 of RP1 resulting in a frame shift and eventually a premature stop codon (p.I263Nfs8*).
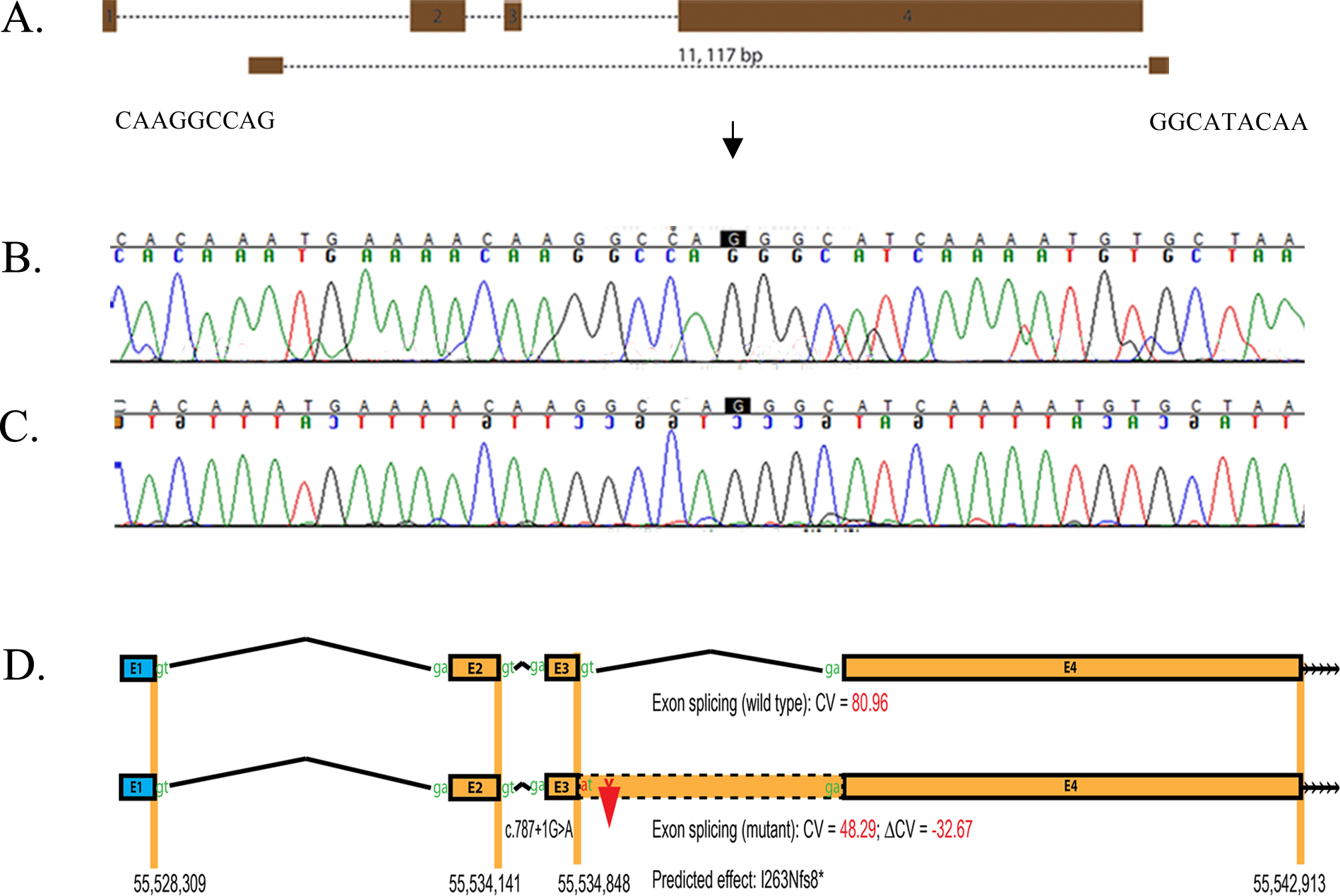
 Figure 5 of
Kabir, Mol Vis 2016; 22:610-625.
Figure 5 of
Kabir, Mol Vis 2016; 22:610-625.  Figure 5 of
Kabir, Mol Vis 2016; 22:610-625.
Figure 5 of
Kabir, Mol Vis 2016; 22:610-625. 