Figure 6. Structural domains in the EPHA2 protein and location of the mutations analyzed in the study. Schematic diagram of the EPHA2
protein showing the ligand binding (brown), fibronectin III repeats (purple), transmembrane segment (light blue), kinase (dark
blue), SAM (red), and PDZ (gray) domains. The helical loops (H1–H5) formed in the secondary structure of the SAM domain are
also shown. The positions of the mutations analyzed in this study are marked. The mutations that mis-localized in MDCK and
Caco-2 cells are highlighted in orange, and those that showed normal localization are highlighted in green. The p.P584L mutation
affects a residue in the juxtamembrane region of the protein, but does not affect protein localization. The mutant EPHA2 proteins
that mis-localize to the perinuclear region carry p.T940I and p.D942fsXC71 mutations that affect helix 4 (H4) in the SAM domain.
The p.A959T mutation alters a residue in helix 5 (H5) in the SAM domain, and the p.V972GfsXC39 mutation affects residues in
the SAM and PDZ domains of EPHA2 but do not affect protein localization.
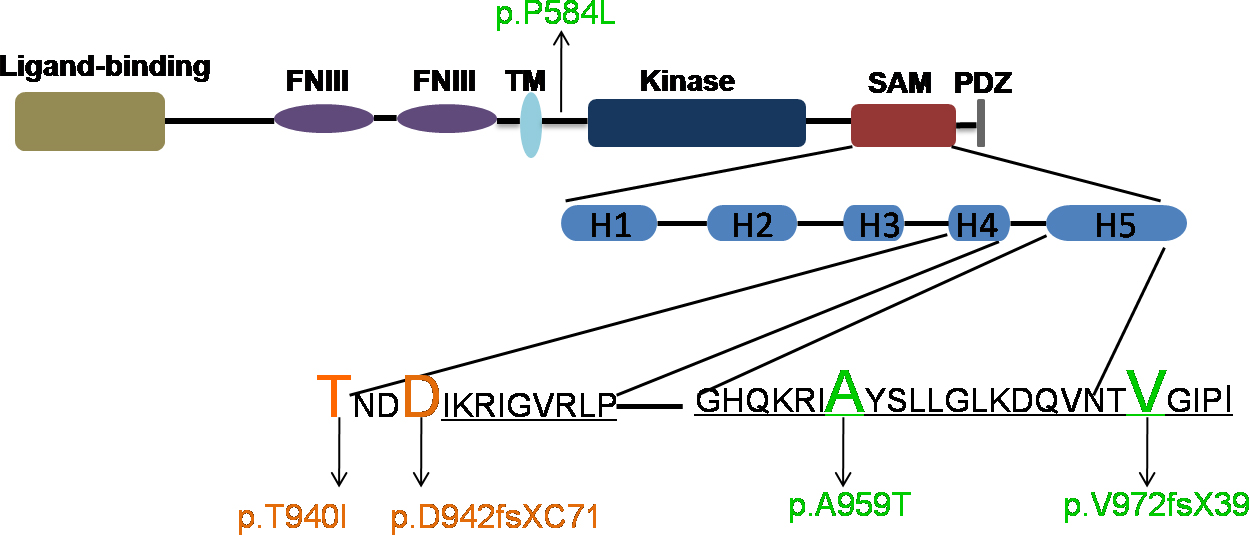
 Figure 6 of
Dave, Mol Vis 2016; 22:18-30.
Figure 6 of
Dave, Mol Vis 2016; 22:18-30.  Figure 6 of
Dave, Mol Vis 2016; 22:18-30.
Figure 6 of
Dave, Mol Vis 2016; 22:18-30. 