Figure 1. Depiction of the PIKFYVE gene with associated functional domains found using PredictProtein. Mutations have been reported in exons 15, 16, 19, 20,
and 24. The novel mutations that we report are indicated in red. Predicted binding sites and secondary structures are depicted
in relation to the PIKFYVE protein. Functional domains are denoted by letters: A, zinc finger phosphoinositide kinase; B,
Disheveld EGL-10, plextrin homology domains; C, β-sheet winged helix DNA/RNA-binding motif; D, cytosolic chaperone CCTγ apical
domain-like motif; E, spectrin repeats; and F, C-terminal fragment similar to the common kinase core found in II β phosphatidylinositol-4-phospohate
5-kinase. Binding regions are designated by shapes: Circles represent polynucleotide-binding regions, and diamonds represent
protein binding regions. Secondary structures are denoted by blue and red boxes: Blue boxes represent beta strands, and red
boxes represent alpha helices.
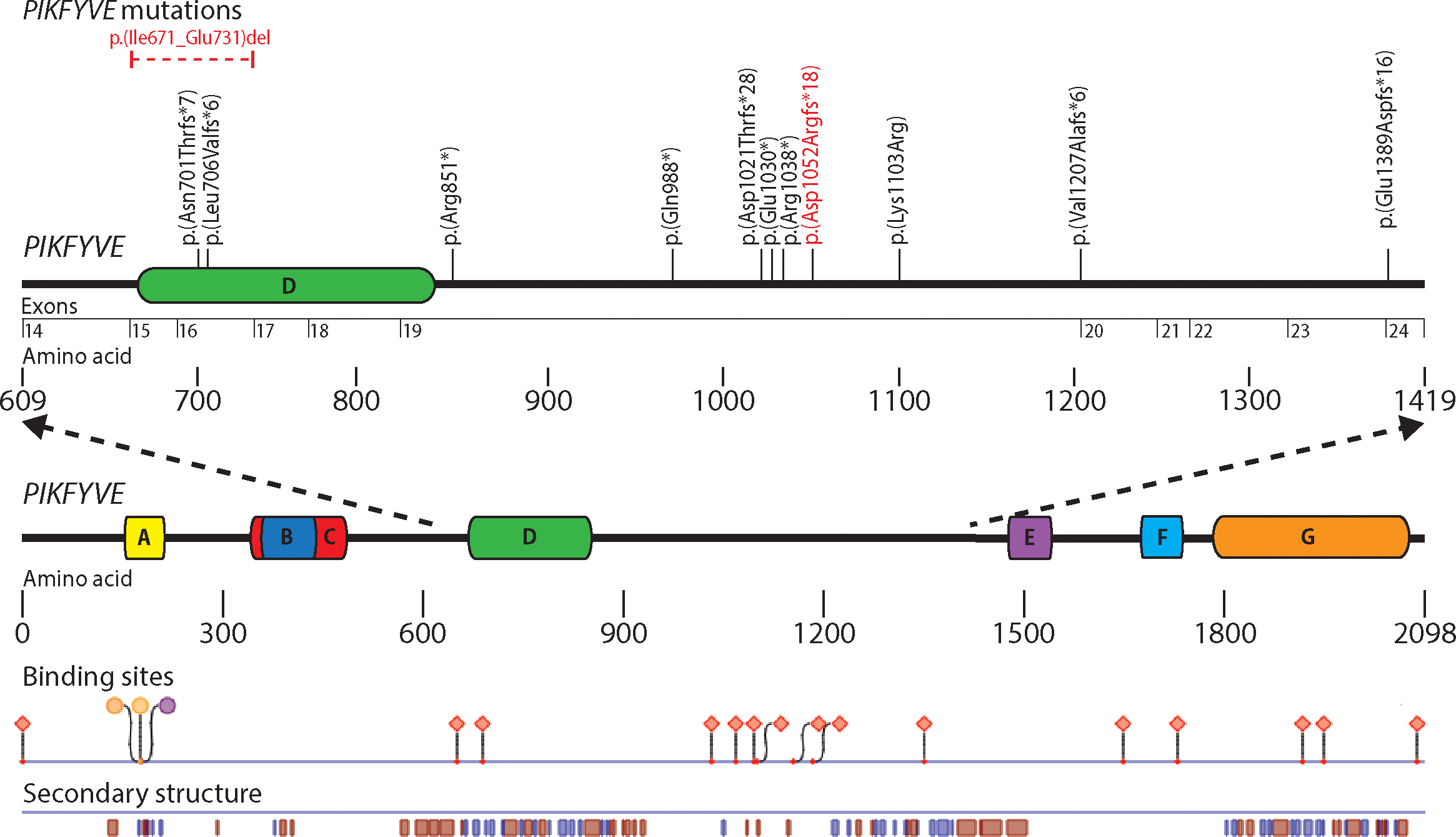
 Figure 1 of
Gee, Mol Vis 2015; 21:1093-1100.
Figure 1 of
Gee, Mol Vis 2015; 21:1093-1100.  Figure 1 of
Gee, Mol Vis 2015; 21:1093-1100.
Figure 1 of
Gee, Mol Vis 2015; 21:1093-1100. 