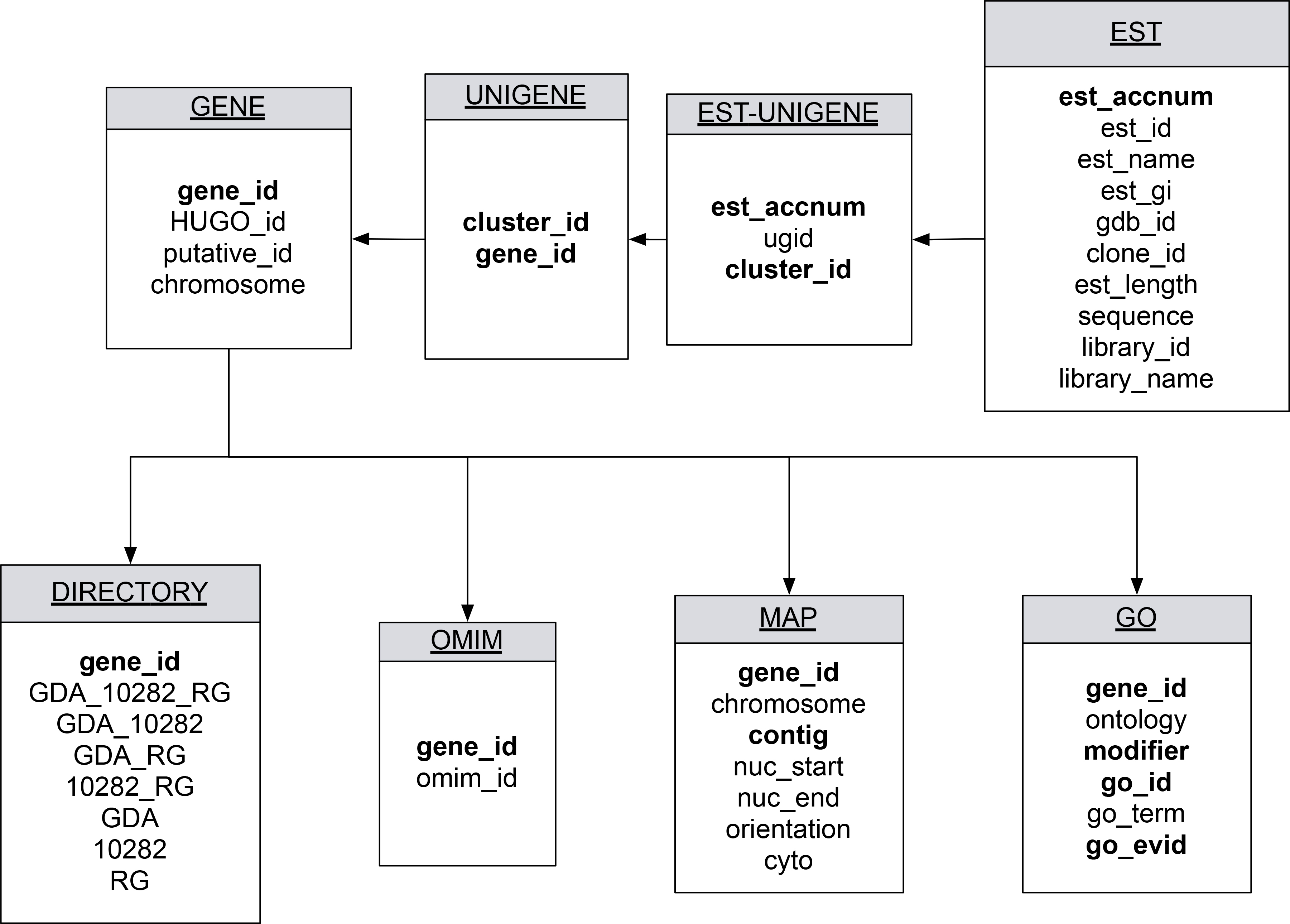
Figure 2. The relational database schema. Each box describes a table, and each line describes a connection between tables. Bolded phrases
within each table indicate key elements used to link between tables. Data for the table expressed sequence tag (EST) is derived
from DataBase of
Expressed Sequence Tags (dbEST); table EST-UNIGENE is derived from UniGene, and all remaining tables are derived from GenedB data.
EST: Table describing data collected from dbEST. This includes GenBank accession number, EST ID, EST name, EST gene identifier
(gi), GDB ID, IMAGE clone ID, length of the EST, sequence of the EST, and both ID number and name of the library from which
the EST is derived.
EST-UNIGENE: This table links the GenBank accession number to the UniGene clusters. The UGID is a novel identifier included for future
expansion.
UNIGENE: This table links the UniGene cluster to the Gene database identifiers. There is not necessarily a one-to-one relationship
between UniGene and Gene.
GENE: This describes data collected from the Gene database. This includes the gene symbol (HUGO ID), gene name, and chromosome
of origin.
GO: This table describes data relevant to Gene Ontology (GO) annotations. Each gene may have 0 or more GO annotations associated
with it; of these annotations, they may have a term modifying their meaning (such as NOT or CONTRIBUTES_TO; “modifier”), a
numerical identifier and text definition (GO ID and GO TERM) as well as all the evidence codes associated with that combination
of modifier and GO annotation.
MAP: This table describes map relevant data. Some genes may map to multiple chromosomes (for example, pseudoautosomal genes)
so allowance is made for multiple map locations for a given gene. Chromosome, contig (useful for genes not fully mapped yet),
nucleotide positions, orientation, and cytogenetic band (cyto) information is included.
OMIM: This data describes OMIM data associated with a given Gene.
DIRECTORY: This complex table tracks which experiment contributed genes to the entire set. Genes that are identified by a single experiment
are listed under “GDA,” “10282” or “RG” (Research Genetics); doubly identified genes by one of the three paired listings,
and triply identified genes by the table GDA_10282_RG.
 Figure 2 of
Ziesel, Mol Vis 2014; 20:947-955.
Figure 2 of
Ziesel, Mol Vis 2014; 20:947-955.  Figure 2 of
Ziesel, Mol Vis 2014; 20:947-955.
Figure 2 of
Ziesel, Mol Vis 2014; 20:947-955.