Figure 1. Transcriptional regulation by PAX6 and PAX6(5a) on P6CON luciferase reporters. A: A schematic diagram of PAX6 and PAX6(5a) proteins including six missense mutations G18W, R26G, N50K, G64V, R128C, and R242T.
Paired domain, PD; homeodomain, HD, proline/serine/threonine-rich domain, PST; β-turn, β; linker between the PAI and RED subdomains,
L. B: Distribution of nucleotides in the P6CON Pax6-binding site. The DNA sequence for gene synthesis is aligned with the P6CON
motif. The binding sites and flanking sequences are in upper and lower case, respectively. C: A schematic representation of the 6x Pax6 DNA-binding motif/E4TATA-luciferase reporter. D: Evaluation of PAX6 (black bars) and PAX6(5a) (gray bars) series in cotransfected P19 cells. The data are expressed as relative
fold changes elicited in the presence of PAX6 or PAX6(5a) compared with the changes found with the empty vector, pKW10. The
sample size n=6 from two independent triplicates. The error bars represent the standard deviation. The significant fold-changes
are indicated by the asterisks. The range of the p values are: *** <0.001, ** 0.001~0.01, * 0.01~0.05. The p values between
empty vector and wild-type Pax6/Pax6(5a) are calculated with paired Student t tests. The p value for each mutant is calculated by comparing it with the wild-type.
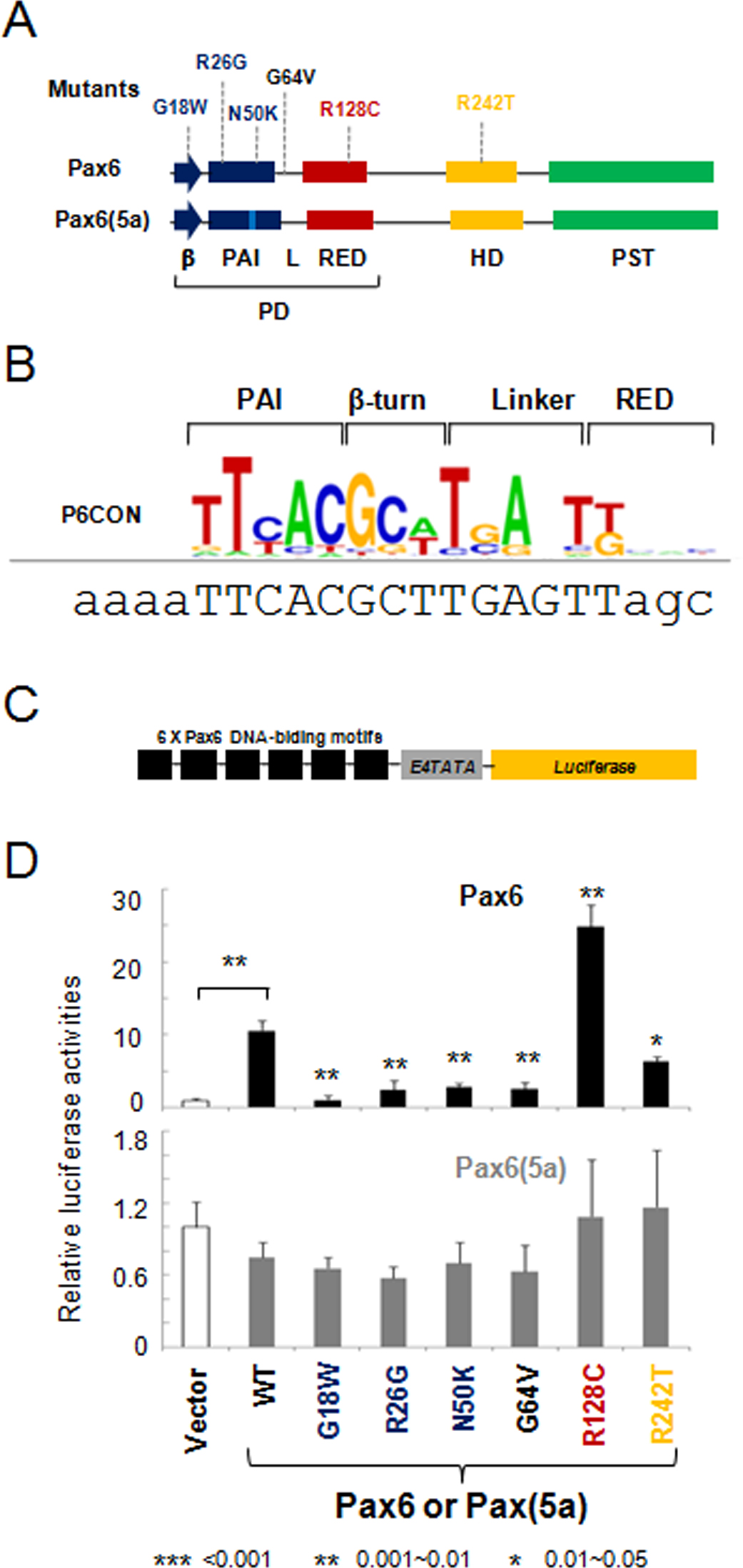
 Figure 1 of
Xie, Mol Vis 2014; 20:270-282.
Figure 1 of
Xie, Mol Vis 2014; 20:270-282.  Figure 1 of
Xie, Mol Vis 2014; 20:270-282.
Figure 1 of
Xie, Mol Vis 2014; 20:270-282. 