Dept. of Cell Biology and Human Anatomy
*corresponding author email: jfhess@ucdavis.edu
School of Medicine
University of California
Davis, CA 95616
Abstract
Clarity of the mammalian lens is due in part to the complete lack of internal organelles, including nuclei, within the lens fiber cells that compose the bulk of the lens. Experimental evidence shows that as the differentiation of lens fiber cells progresses, nuclei and nuclear DNA are actively degraded. Prior characterization of chick lens development suggests that DNase I could be involved in lens DNA degredation. However, recent data suggest that DNase I is unlikely to be the nuclease responsible for DNA degredation in the differentiating lens. In this report, we find that in the murine lens, mRNA for DNase I is undetectable by northern blotting or PCR. We conclude that mRNA for DNase I is either not present or present in very low levels in murine lens. Our results are consistent with the hypothesis that DNase I is not involved in lens DNA degredation.
Introduction
The differentiation of a lens epithelial cell into an elongated lens fiber cell is a terminal process that results in the elimination of cellular organelles, including the nucleus and its DNA (1, 2). With regard to the degradation of cellular DNA, evidence from the developing avian lens supports the hypothesis that DNA degradation is an active process (3-8). The enzymes responsible for the lens DNA degradation have not yet been identified. Thus, the differentiation of a lens fiber cell includes a process (DNA degradation) that may be related to the end stages of apoptotic cell death.
From studies of apoptosis, two different DNase enzymes have been suggested to be responsible for DNA degradation at the end of apoptotic cell death (9, 10). DNase I, which has been cloned and sequenced from the rat (11), human (12), and mouse (10) has been shown to be expressed in thymocytes, and is strongly suggested to be the nuclease responsible for DNA degradation in negative selection of immature thymocytes (13). Functionally, the endonuclease activated during thymocyte apoptosis requires Ca2+/Mg2+ and is inhibited by Zn2+; activity is optimal at neutral pH (14-16). In contrast, characterization of apoptosis induced in CHO cells by either cytotoxic agents or hyperthermia implicates DNase II as the enzyme responsible for DNA degradation (17). Functionally, DNase II activity is independent of added Ca2+/Mg2+, with a pH optimum of ~5. In addition to differences in pH optima and divalent cation requirements, DNase I and DNase II produce different 5' and 3' termini. DNase I produces DNA oligomers with 5' phosphates and 3' hydroxyls while DNase II produces 3' phosphates and 5' hydroxyls.
Evidence from chick lens extracts indicates that two distinct DNase activities are present in lens fiber cells (5-6). One would appear to be similar, if not identical to DNase I: ~30 kD as estimated by SDS-PAGE, requires Ca2+/Mg2+, and is active at neutral pH (6). The second, ~40 kD by SDS-PAGE, is inhibited by millimolar Ca2+/Mg2+ but is also inhibited by EDTA, indicating that trace amounts of divalent cations are required (6).
We report here efforts to more explicitly define the nature of the DNase activity in lens fiber cells by using PCR and northern blotting to probe for DNase I sequences. The results suggest that either the mRNA for DNase I is of very low abundance and thus undetectable by these methods, or DNase I is not expressed in the mouse lens fiber cell during differentiation. The latter possibility seems more likely and is consistent with published results (8, 9) that show that 3' hydroxyls are not formed during lens DNA degradation and DNase II is involved in nuclear degradation during lens cell differentiation.
Materials and Methods
Total RNA was isolated from mouse kidney and lens by the acid-phenol-guanidinium isothiocyanate procedure (18) using 21 day old Swiss-Webster mice purchased from Simonson Laboratories (Gilroy, CA). Total lens RNA was isolated from decapsulated mouse lenses by incubation in the GITC solution to dissolve the outer lens fiber cells; the inner nucleus as well as older fiber cells adherring to the nucleus were discarded. Gel electrophoresis of RNA in formaldehyde-MOPS agarose gels was as described in Sambrook, et. al. (19). Agarose gels were transferred to nylon membranes (MSI, Westboro, MA) by capillary action, followed by UV cross linking. Northern blotting was performed by hybridization in 50% formamide, 5X SSPE, 5X Denhardts (20), and 0.5% SDS. Filters were washed (2X, 15 minutes each) at room temperature in 2X SSPE, 0.1% SDS followed by high stringency washing for 15 minutes at 60°C in 0.2X SSPE, 0.1% SDS. Radioactive probes were prepared with the Oligolabelling kit (Pharmacia, Piscataway, NJ). When required, filters were stripped by boiling in 0.1% SDS for 5 minutes.
Reverse transcription of total RNA was performed with either random hexamer or dT containing primers (9, 20). Oligonucleotides 2749 (5'- CATATGCTGAGAATTGCAGCCTTCAACA-3'[corresponding to nucleotides 261-282 of genbank sequence U00478 {mouse DNAse I}]) and 2750 (5'-TCTGAGTGTCACCTCCACTGGGTAATG-3' [corresponding to nucleotides 2475-2501 of Genbank sequence U00478]), were used to amplify the coding region of DNase I corresponding to the mature protein (13, 21). Amplifications were performed using conditions specified in Barry and Eastman (10). The amplified kidney DNase I product was purified on a low-melting temperature agarose gel followed by purification using Magic PCR preps (Promega Biotech, Madison, WI) and cloned into Sma I- digested pSP72 using the Sureclone kit (Pharmacia, Piscataway, NJ). To verify the authenticity of the PCR amplified DNase I sequence, DNA sequencing was performed using the Taquence DNA sequencing kit (USB, Indianapolis, IN) and sequences were analyzed using MacDNAsis software (Hitachi Software, Pleasanton, CA). Additional mouse-specific oligonucleotide primers were synthesized by the UC Davis Protein Structure Laboratory. Oligonucleotides 3560 (5'-CTAGGTAAAATCGTAGAGGG corresponding to nucleotides 1654-1674 of Genbank entry U00478) and 2749 (above) were used to amplify a 470 bp region of mouse DNase I cDNA. As a positive control for lens RNA integrity, amplification of mouse CP49, a lens fiber cell specific cytoskeletal protein, was performed using oligonucleotides 1773 (5'-GATCGTCATATGAGCAAGAGGAGAGTGGCAGCGGACTTG) and 1700 (5'-CCCGCAGTTGTGTTTCCAGTTCC) (Hess, Casselman and FitzGerald, unpublished); the amplified product is 442 bp.
Results
To maximize the possibility of detecting mouse lens DNase sequence, we created homologous primers and probes from mouse kidney DNase I sequence. Pietsch, et. al., in their paper describing rat DNase I, identified conditions that were successful at amplification of DNase I sequences from mouse kidney RNA samples (13). We therefore isolated total RNA from mouse kidney and made total cDNA pools using reverse transcriptase and either random hexamer or dT-adapter primers (20). Amplification of DNase I sequences from mouse kidney produced the predicted ~800bp signal. Verification of the authenticity of this PCR product as mouse DNase I was obtained by determining the DNA sequence of the fragment and northern blotting. Efforts to amplify DNase I activity in cDNA made from mouse lens were consistently negative.
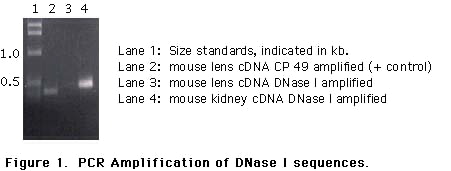
To confirm this result, additional amplification reactions were designed using one of the original amplification primers (2749) and an second primer initially ordered for DNA sequencing (3560). Our hope was that amplification of a smaller region might be successful when a larger region, for unknown reasons was not. Using oligonucleotides designed to amplify a 470 bp region of mouse DNase I, we established that kidney cDNA was positive for amplification while lens cDNA was again negative. Figure 1 shows a typical example: lane 4 is a mouse kidney cDNA sample; lane 3 is the same reaction performed with lens cDNA input. Lane 2 shows a lens cDNA positive control, amplification of mouse CP49 sequences (16). Lane 1 is the Life Technologies 1 kb ladder. Amplification of cDNA samples from both random hexamer and dT-primed cDNA pools failed to yield the expected signals (data not shown). These data suggest RNA sequences for mouse DNase I are not present in lens, or present in extremely low copy number.

To demonstrate that the negative results were not due to RNA degradation, we electrophoresed samples of the mouse lens and kidney RNA on agarose gels. Ethidium-bromide staining showed no obvious degradation, and the gel was transferred to a nylon membrane (Figure 2, panel A). Hybridization of the membrane with a radioactively-labeled mouse DNase I probe revealed hybridization only to the kidney RNA (Figure 2, panel B, lane K). Based on migration of RNA standards electrophoresed in parallel, the positive signal is ~1.6-1.8 kb. After autoradiography, the membrane was stripped and probed with a mouse CP49 probe which is specific for lens fiber cell RNA (16). As can be seen in Figure 2, panel C, no hybridization of the CP49 probe is seen in the mouse kidney lane "K", while an abundant signal is present in the lens RNA sample. The signal in the mouse lens RNA lane is consistent with previous data (Hess, Casselman and FitzGerald, unpublished). Thus, conditions that produce a vigorously positive response from mouse kidney produce no detectable signal from lens.
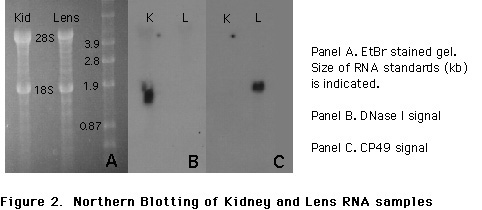
The signal present in the center panel verifies results obtained from RT-PCR: mouse kidney RNA contains DNase I-encoding sequences and mouse lens RNA does not. In panel C, the signal produced in the lens RNA lane in the autoradiography on the right clearly shows that the lens RNA is intact and undegraded. Thus, any DNase I sequences should have given a positive signal if present. The results of the DNase I and CP49 probing of the same filter are completely concordant with the results of PCR.
Discussion
In an effort to define the molecular mechanisms responsible for DNA degradation which occurs during lens fiber cell differentiation, a process that bears partial resemblance to apoptosis, we probed mouse lenses for evidence of DNase I sequence. We isolated mouse cDNA sequences and used two different techniques to analyze mouse lens RNA for the presence of DNase I sequences. From the results of both PCR and northern blotting, we report that DNase I mRNA sequences are not detectable in the mouse lens.
The degradation of the nucleus and all organelles in the lens fiber cell has been known for many years. Assumed, but only recently investigated, was the hypothesis that nuclear DNA degradation and removal is an active process requiring expression of a particular set of genes, including nucleases, similar in part to apoptosis. Indeed, recent studies have succeeded in identifying nuclease activities present in the lens during chick lens development (4). Subsequent characterization provided evidence that two distinct nuclease activities were present, each with a set of functional requirements that were consistent with the nucleases DNase I and DNase II.
To examine the hypothesis that DNase I is present in the lens, we have used the DNA sequence available for DNase I (11, 13) to isolate mouse DNase I, and then probed mouse lens RNA samples for the DNase. Our initial attempts to amplify DNase I from lens samples were negative; a representative example is shown along with a positive control amplification of CP49 cDNA sequences (see Figure 1). Alteration of amplification conditions did not lead to amplification of DNase sequences from lens RNA (data not shown).
To further investigate the possibility that low levels of DNase I mRNA are present, we used northern blotting to probe lens and kidney samples. As shown in Figure 2, no signal for DNase I mRNA sequences is apparent in the lens sample under conditions which give a strong signal in the kidney RNA sample (panel B). Probing of the same filter for CP49, a fiber cell-specific RNA, reveals that the lens RNA sample is not significantly degraded, reducing the possibility that the negative results were caused by RNA degradation. Thus, two nucleic acid-based approaches to demonstrate the presence of DNase I mRNA in the lens have produced negative results.
The lack of readily detectable DNase I mRNA sequences in the lens is surprising. On one hand, enzymatic characterizations provide preliminary evidence for DNase I (or DNase I-like activity) in the lens (6). However, mechanistic evidence has also been presented that argues against the presence of DNase I in the lens: 3' hydroxyls are not produced by nuclease activities present in the developing avian lens. The generation of 3' hydroxyls would be expected if DNase I were the enzyme responsible for the observed degradation (8). That we could not readily detect DNase I mRNA sequences is of course not definitive evidence that DNase I is not present; however, our data indicating a lack of DNase I mRNA is consistent with data indicating DNase I cleavage products are not present in the lens.
As for the possibility that DNase II is the enzyme responsible for DNA degradation in the lens, additional experiments are necessary. Although amino acid sequence data from the active site of DNase II has been published (22), to the best of our knowledge, no nucleic acid-based clones have been reported. Thus, examination of lens RNA samples for DNase II mRNA by nucleic acid-based methods is not possible at this time. However, examination of the lens for DNase II protein has been performed by Torriglia et al. (9) and they report that DNase II is likely the enzyme responsible for lens fiber cell degradation.
An additional enzymatic activity potentially responsible or partially responsible for lens DNA degradation could be an endo-exonuclease with both single- and double-stranded nuclease activities (23, 24). Enzymatically, this nuclease requires a divalent cation and is most active at pH 7.5 (ds-DNase activity) and pH 8.0 (ss-DNase activity). As with DNase II, nucleic acid probes are unavailable at this time. Thus, despite the reports that DNase activity is present in the lens, our efforts to detect DNase I sequences in lens cDNA were consistently negative. This suggests that the DNase I activity reported in lens derives from an enzyme other than the DNase I, or that DNase I is present, but its message exists in extremely low abundance, below the detection limits of the technique employed here.
References
1. Bloemendal H. Molecular and Cellular Biology of the Ocular Lens. New York, New York: J. Wiley and sons, 1981.
2. Maisel H. The Ocular Lens. New York, New York: Marcel Dekker, 1985.
3. Yamamoto A, Araki T, Counis MF. Decrease of DNA per cell during development of the lens in chickens. Histochemistry 1990;94(3):293-6. ![]()
4. Counis MF, Chaudun E, Courtois Y, Allinquant B. Lens fiber differentiation correlated with activation of two different DNAases in lens embryonic cells. Cell Differ Dev 1989;27(2):137-46. ![]()
5. Counis MF, Chaudun E, Allinquant B, et al. The lens: a model for chromatin degradation studies in terminally differentiating cells. Int J Biochem 1989;21(3):235-42. ![]()
6. Counis MF, Chaudun E, Courtois Y, Allinquant B. DNAase activities in embryonic chicken lens: in epithelial cells or in differentiating fibers where chromatin is progressively cleaved. Biol Cell 1991;72(3):231-8. ![]()
7. Appleby DW, Modak SP. DNA degradation in terminally differentiating lens fiber cells from chick embryos. Proc Natl Acad Sci U S A 1977;74(12):5579-83. ![]()
8. Chaudun E, Arruti C, Courtois Y, et al. DNA strand breakage during physiological apoptosis of the embryonic chick lens: free 3' OH end single strand breaks do not accumulate even in the presence of a cation-independent deoxyribonuclease. J Cell Physiol 1994;158(2):354-64. ![]()
9. Torriglia, A., Chaundun, E., Chany-Fournier, F., Jeanny, J.-C., Courtois, Y., and M.-F. Counis. Involvement of DNase II in nuclear degeneration during lens cell differentiation. J. Biol. Chem. 1995: 270(48):28579-28585. ![]()
10. Barry MA, Eastman A. Identification of deoxyribonuclease II as an endonuclease involved in apoptosis. Arch Biochem Biophys 1993;300(1):440-50. ![]()
11. Peitsch MC, Irmler M, French LE, Tschopp J. Genomic organisation and expression of mouse deoxyribonuclease I. Biochem Biophys Res Commun 1995;207(1):62-8. ![]()
12. Stasiak PC, Lane EB. Sequence of cDNA coding for human keratin 19. Nucleic Acids Res 1987;15(23):10058. ![]()
13. Shak S, Capon DJ, Hellmiss R, Marsters SA, Baker CL. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc Natl Acad Sci U S A 1990;87(23):9188-92. ![]()
14. Peitsch MC, Polzar B, Stephan H, et al. Characterization of the endogenous deoxyribonuclease involved in nuclear DNA degradation during apoptosis (programmed cell death). Embo J 1993;12(1):371-7. ![]()
15. Romano V, Raimondi E, Bosco P, et al. Chromosomal mapping of human cytokeratin 13 gene (KRT13). Genomics 1992;14(2):495-7. ![]()
16. Cohen JJ, Duke RC. Glucocorticoid activation of a calcium-dependent endonuclease in thymocyte nuclei leads to cell death. J Immunol 1984;132(1):38-42. ![]()
17. Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 1980;284(5756):555-6. ![]()
18. Modak SP, Perdue SW. Terminal lens cell differentiation. I. Histological and microspectrophotometric analysis of nuclear degeneration. Exp Cell Res 1970;59(1):43-56. ![]()
19. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987;162(1):156-9. ![]()
20. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. (second edition ed.) Cold Spring Harbor, New York: Cold Spring Harbor Press, 1989.
21. Glaser T, Lane J, Housman D. A mouse model of the aniridia-Wilms tumor deletion syndrome. Science 1990;250(4982):823-7. ![]()
22. Paudel HK, Liao TH. Comparison of the three primary structures of deoxyribonuclease isolated from bovine, ovine, and porcine pancreas. Derivation of the amino acid sequence of ovine DNase and revision of the previously published amino acid sequence of bovine DNase. J Biol Chem 1986;261(34):16012-7. ![]()
23. Liao TH. The subunit structure and active site sequence of porcine spleen deoxyribonuclease. J Biol Chem 1985;260(19):10708-13. ![]()
24. Couture C, Chow TY. Purification and characterization of a mammalian endo-exonuclease. Nucleic Acids Res 1992;20(16):4355-61. ![]()
25. Fraser MJ. Endo-exonucleases: enzymes involved in DNA repair and cell death? Bioessays 1994;16(10):761-6. ![]()