A Molecular Vision Short Report
The gene for PEDF, a retinal growth factor, is a prime candidate for retinitis pigmentosa
and is tightly linked to the RP13 locus on chromosome 17p13.3
Rene Goliath1, Joyce Tombran-Tink2, Ignacio R. Rodriquez2, Gerald Chader2, Rajkumar
Ramesar1 and Jacquie Greenberg1*
1Department of Human Genetics, MRC Research Unit of Medical Genetics, University of
Cape Town Medical School Observatory, Cape Town, South Africa and 2Laboratory
of Retinal Cell and Molecular Biology, National Eye Institute, NIH, USA
*To whom correspondence should be addressed (Email: jg@anat.uct.ac.za)
Pigment epithelium-derived factor (PEDF), a unique 50 kDa protein secreted by human
fetal retinal pigment epithelial cells, has been shown to have neurotrophic effects
on human Y-79 retinoblastoma cells in vitro. The gene for PEDF is highly expressed by both fetal human and young adult retinal pigment epithelium (RPE) cells
and is down-regulated in senescent but not quiescent RPE cells (5).
The cDNA for the human PEDF has been mapped to the short arm of chromosome 17 (17p13-ter)
(4) and an autosomal dominant Retinitis Pigmentosa (RP13) gene locus has been assigned
to the same chromosomal region in a large South African family (2). Here we refine the physical mapping of the PEDF gene and provide evidence for a more distal
localization (17p13.3) with respect to the previous report (4).
In the mapping of the autosomal dominant RP (ADRP) locus, a series of three point analyses was undertaken
with two markers and the disease gene. A peak lod score was obtained between D17S849
and D17S938 indicating the most likely interval containing this ADRP gene (1). Five additional markers between D17S849 and D17S938 were genotyped in order to further
refine the region containing this ADRP-causing gene. Another series of multipoint
analyses were performed and the marker D17S1528 showed tight linkage to the disease
locus with no recombinations (Z = 7.19). Two markers, D17S1529 and D17S831, located on
either side of D17S1528, showed significant positive lod scores of 5.64 and 6.66,
at theta = 0.05. The microsatellite markers used have been ordered as follows (Genethon:
unpublished data):
cen - D17S796 - D17S938 - D17S1353 - D17S829 - D17S831 - D17S1528 - D17S1529 - D17S849 - D17S926 - tel.
ADRP-SA1 has been placed in the interval between D17S1529-D17S831, while D17S1529
and D17S831 themselves were excluded. In the study, two-point and multipoint analyses were performed using the Linkage package, version 5.2, run under OS/2 (3).
A gene frequency of 0.001 was assumed for the disease.
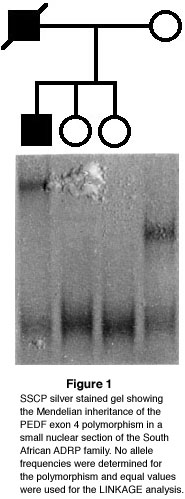 The finding of a polymorphism in exon 4 of the PEDF gene enabled us to genotype this
family with PEDF. This polymorphism was detected on single-stranded conformational
polymorphism (SSCP) analysis. The resulting SSCP pattern facilitated segregation
analysis in the family (Fig.1). Direct sequencing of the PCR products revealed a T to C and
a C to G substitution in codons 130 and 132, resulting in a neutral and a missense
mutation, respectively. The intronic primers used to amplify exon 4 of the PEDF gene
were: 4F 5'-TGAGTATAGTGTCTGTGTTCTGGGA-3' and 4R 5'-AAGACCCCCCCAGCCTGCAGCATGG-3'.
The finding of a polymorphism in exon 4 of the PEDF gene enabled us to genotype this
family with PEDF. This polymorphism was detected on single-stranded conformational
polymorphism (SSCP) analysis. The resulting SSCP pattern facilitated segregation
analysis in the family (Fig.1). Direct sequencing of the PCR products revealed a T to C and
a C to G substitution in codons 130 and 132, resulting in a neutral and a missense
mutation, respectively. The intronic primers used to amplify exon 4 of the PEDF gene
were: 4F 5'-TGAGTATAGTGTCTGTGTTCTGGGA-3' and 4R 5'-AAGACCCCCCCAGCCTGCAGCATGG-3'.
PCR reactions were performed using 200 ng genomic DNA, 200 ÁM of each dATP, dGTP, dCTP
and dTTP, 100 ng of each primer, 50 mM KCl, 10mM Tris HCl pH 8.4, 1.5 mM MgCl2 and 1 unit of Taq DNA polymerase in a total volume of 25 Ál. Cycling conditions were
one cycle at 95°C denaturation for 3 minutes, followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 62°C for 30 seconds, elongation at 72°C for 40 seconds, followed by one extended elongation cycle at 72°C for 5 minutes. For SSCP analysis, the PCR products were diluted 1 to 1 in a loading
buffer (7M urea, 50% formamide, 20 mM Tris-HCl and 10 mM EDTA). The samples were denatured
and loaded directly onto a 0.5x MDE (Hydrolink) gel containing 0.5x glycerol and
0.5x TBE. Gels were electrophoresed at 6 watts constant power overnight and visualised
with routine silver staining.
Two-point linkage analysis between the disease phenotype and this polymorphism produced
a maximum lod score of 7.1 at a theta of 0.00. Subsequent multipoint linkage analysis
allowed us to map the PEDF gene with respect to the tightly linked markers on the short arm of chromosome 17, used in the fine mapping study (1). LINKMAP was used
to perform a four-point linkage analysis where PEDF was moved up to and in between
the 3 fixed loci D17S849, D17S1529 and D17S831. The estimated theta value between
these new markers is 0.03 (Genethon, unpublished data). A theta value of 0.5 was used as
the starting point away from D17S849 and then 4 equal increment values were used
to move in on the fixed marker loci. The pooled results are presented in Figure
2.
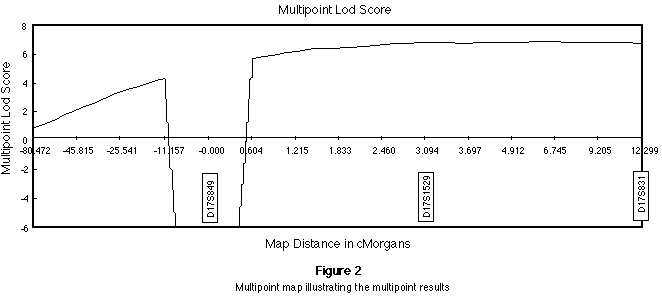 The maximum estimate for the location of PEDF was close to D17S1529 with a maximum
lod score of 6.71. Two point analyses between PEDF and D17S1529, D17S1528 and D17S831
produced maximum lod scores of 6.53, 6.26 and 5.36 respectively, each at a recombination fraction of zero.
The maximum estimate for the location of PEDF was close to D17S1529 with a maximum
lod score of 6.71. Two point analyses between PEDF and D17S1529, D17S1528 and D17S831
produced maximum lod scores of 6.53, 6.26 and 5.36 respectively, each at a recombination fraction of zero.
In conclusion, these results indicate a more distal localization of the PEDF gene
on the short arm of chromosome 17 with respect to previously fine mapped genetic
markers (1). Taken together, these data provide evidence for the localization of
the PEDF gene to the 17p13.3 chromosomal region, distal to the recoverin gene locus at 17p13.1
and very close to the RP13 locus. This result confirms the in situ
sublocalization of PEDF to 17p13.1-pter (4) and places the gene on a more distal
and possibly more confined physical location, very close to D17S1529 on 17p13.3.
The highly informative internal polymorphism in exon 4 of the PEDF gene reported here
could facilitate further candidate gene linkage studies in families with inherited
retinal degeneration. As PEDF is a prime candidate for RP, it is possible that it
is the disease-causing gene in the South African RP family localized to 17p13.3. This
is currently being investigated.
Acknowledgements
This research was supported by grants from the Retinal Preservation Foundation of
South Africa, the South African Medical Research Council, the UCT staff research
fund, the Mauerberger Foundation, and grants from NIH-EY07961. We thank Jon Moller
for performing the linkage analysis and the RP family members for their participation in the
study.
References
1. Goliath, R, Shugart, YY, Janssens, P, Weissenbach, J, Beighton, P, Ramesar, R,
and Greenberg, J, Fine localization of the locus for autosomal dominant retinitis
pigmentosa on chromosome 17p, Am. J. Hum. Genet.
57 (1995) 962-965. 
2. Greenberg, J, Goliath, RG, Beighton, P, and Ramesar, R, A new locus for autosomal
dominant retinitis pigmentosa on the short arm of chromosome 17, Hum. Mol. Genet.
3 (1994) 915-918. 
3. Lathrop, GM, and Lalouel, JM, Easy calculations of Lod scores and genetic risks
on small computers, Am. J. Hum. Genet. 36 (1984) 460-465.
4. Tombran-Tink, J, Pawar, H, Swaroop, A, Rodriquez, I, and Chader, C, Localization
of the gene for Pigment Epithelium-Derived Factor (PEDF) to Chromosome 17p13.1 and
expression in cultured human retinoblastoma cells, Genomics
19 (1994) 266-272.
5. Tombran-Tink, J, Shivaram, SM, Chader, GJ, Johnson, LV, and Bok, D, Expression,
secretion and age-related down regulation of pigment epithelium-derived factor,
a serpin with neurotrophic activity, Neuroscience
15 (1995) 4992-5003.
Received 29 February 1996 | Revised 29 March 1996 | Accepted 14 May 1996 | Uploaded 19 June 1996
Referencing Note: This article may be referenced as: Mol. Vis. 2:5, 1996.
Alternatively, this article may be referenced by its unique URL:
http://www.emory.edu/molvis/v2/goliath
© 1996 Molecular Vision
 The finding of a polymorphism in exon 4 of the PEDF gene enabled us to genotype this
family with PEDF. This polymorphism was detected on single-stranded conformational
polymorphism (SSCP) analysis. The resulting SSCP pattern facilitated segregation
analysis in the family (Fig.1). Direct sequencing of the PCR products revealed a T to C and
a C to G substitution in codons 130 and 132, resulting in a neutral and a missense
mutation, respectively. The intronic primers used to amplify exon 4 of the PEDF gene
were: 4F 5'-TGAGTATAGTGTCTGTGTTCTGGGA-3' and 4R 5'-AAGACCCCCCCAGCCTGCAGCATGG-3'.
The finding of a polymorphism in exon 4 of the PEDF gene enabled us to genotype this
family with PEDF. This polymorphism was detected on single-stranded conformational
polymorphism (SSCP) analysis. The resulting SSCP pattern facilitated segregation
analysis in the family (Fig.1). Direct sequencing of the PCR products revealed a T to C and
a C to G substitution in codons 130 and 132, resulting in a neutral and a missense
mutation, respectively. The intronic primers used to amplify exon 4 of the PEDF gene
were: 4F 5'-TGAGTATAGTGTCTGTGTTCTGGGA-3' and 4R 5'-AAGACCCCCCCAGCCTGCAGCATGG-3'.
 The maximum estimate for the location of PEDF was close to D17S1529 with a maximum
lod score of 6.71. Two point analyses between PEDF and D17S1529, D17S1528 and D17S831
produced maximum lod scores of 6.53, 6.26 and 5.36 respectively, each at a recombination fraction of zero.
The maximum estimate for the location of PEDF was close to D17S1529 with a maximum
lod score of 6.71. Two point analyses between PEDF and D17S1529, D17S1528 and D17S831
produced maximum lod scores of 6.53, 6.26 and 5.36 respectively, each at a recombination fraction of zero.