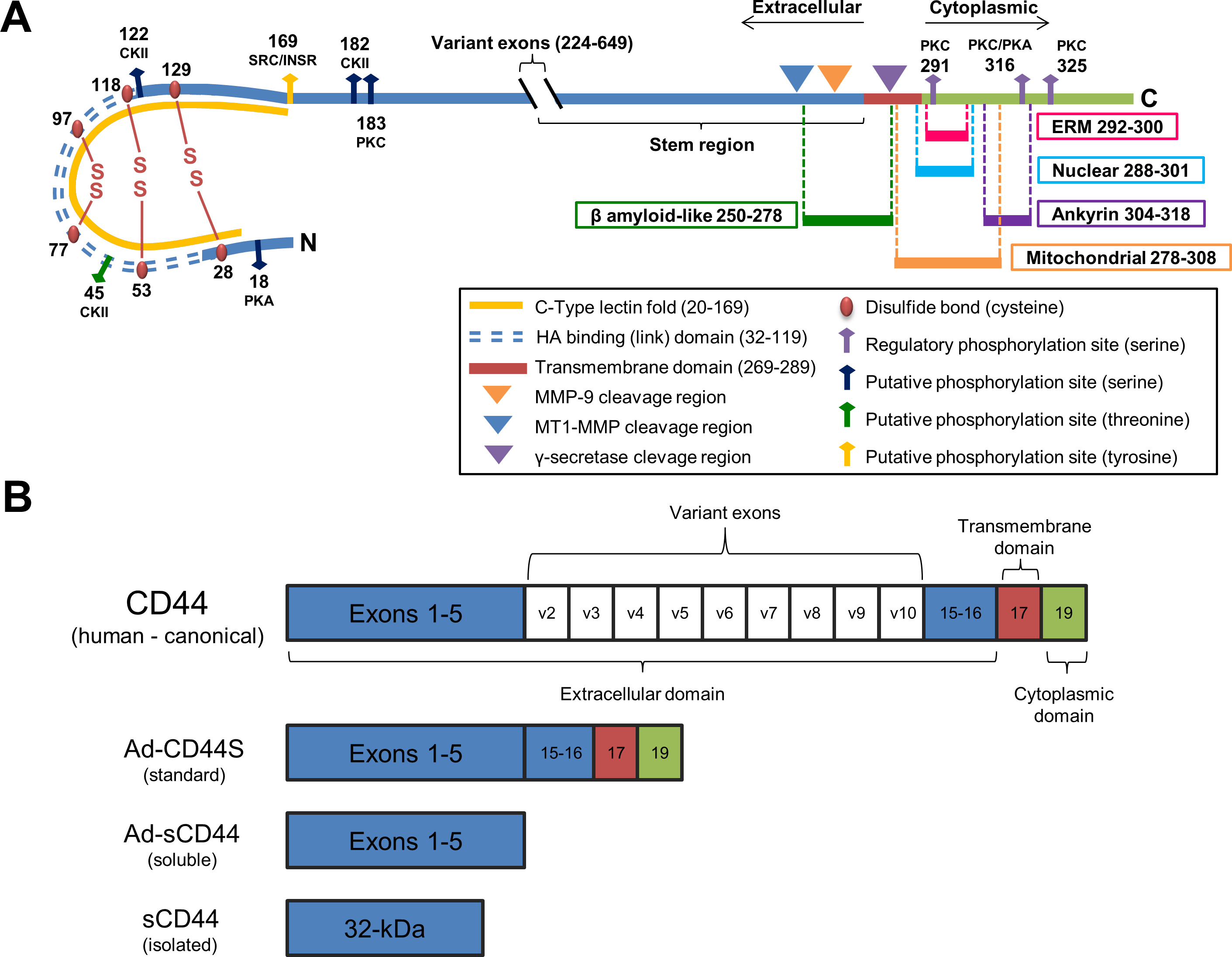
Figure 1. Schematic representation of CD44S.
A: The extracellular (blue), transmembrane (red), and cytoplasmic (green) domains are illustrated for standard CD44S. The extracellular
domain is characterized by cysteine residues that form disulfide bonds (maroon beads), including the two signature disulfide
bonds that form the backbone of the link domain. Several extracellular putative phosphorylation sites as well as docking sites
for MMP-9 and cleavages of CD44 by MT1-MMP (in conjunction with ADAMs 10 and 17) in the CD44 stem region and the transmembrane
region by γ-secretase are shown. The cytoplasmic domain is characterized by three cytoplasmic phosphorylation sites (purple
arrows) that modulate CD44 ezrin linking [
46,
47]. Note the extracellular β amyloid-like fragment and the nuclear and mitochondrial trafficking signals. Abbreviations for
putative phosphorylation sites are: PKA, protein kinase A; PKC, protein kinase C; CKII, casein kinase II; SRC, sarcoma tyrosine
kinase; INSR, insulin receptor kinase; ERM, ezrin, radixin, moesin.
B: Gene structure of canonical CD44 with invariant exons 1–5, variable spliced exons 6–14, invariant exons 15–16 forming stem
region, transmembrane exon 17, and cytoplasmic domain exon 19 is illustrated. Note exon 18 is spliced out [
48]. Adenoviral constructs were the standard (Ad-CD44S) and truncated soluble (Ad-sCD44) isoforms; the isolated sCD44 is depicted
for comparison to the constructs.
 Figure 1 of
Giovingo, Mol Vis 2013; 19:2151-2164.
Figure 1 of
Giovingo, Mol Vis 2013; 19:2151-2164.  Figure 1 of
Giovingo, Mol Vis 2013; 19:2151-2164.
Figure 1 of
Giovingo, Mol Vis 2013; 19:2151-2164.