Figure 3. Alignment of the in vivo cleavage sites in transforming growth factor beta induced protein (TGFBIp) from the lattice corneal
dystrophy (LCD) type 1 variant and healthy corneas. The sequences of the non-tryptic cleavage sites in TGFBIp from the amyloid
deposits (A), periamyloid corneal tissue (B), and the healthy corneal stroma (C) were aligned (from position −5 to +5). Residues with aromatic side chains (F, Y, W, H) and hydrophobic side chains (A, I,
L, M, F, W, Y, V) are marked with red bold letters and gray background, respectively. The residue numbers in TGFBIp of the
aligned sequences are indicated, and the black arrows show the cleavage sites. The position of the amyloidogenic fourth fasciclin
1 (FAS1-4) domain (residues 505–632) is indicated in the alignments. (A) The TGFBIp FAS1–4 domain from the amyloid deposits is predominantly cleaved at the C-termini of the hydrophobic residues
leucine and alanine. (B) All proteolytic cleavages in the TGFBIp FAS1–4 domain from the periamyloid tissue from the LCD type 1 variant cornea occur
at the C-termini of the aromatic side chains (F, Y) and the hydrophobic side chains (A, L). (C) The two in vivo cleavages detected in the TGFBIp FAS1–4 domain from the healthy cornea were at the C-termini of the aromatic
residues phenylalanine and tyrosine. Sequences shown in bold italics indicate cleavage sites, which are also identified in
the healthy cornea.
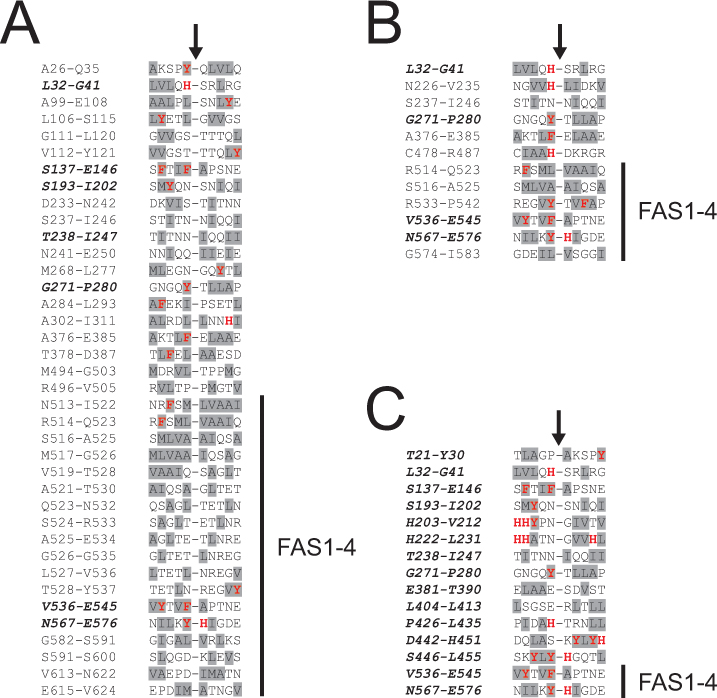
 Figure 3 of
Karring, Mol Vis 2013; 19:861-876.
Figure 3 of
Karring, Mol Vis 2013; 19:861-876.  Figure 3 of
Karring, Mol Vis 2013; 19:861-876.
Figure 3 of
Karring, Mol Vis 2013; 19:861-876. 