Figure 1. CRYAA and CRYGC mutations in two Korean families. A: Pedigrees of families A and B are indicated. Black and open symbols denote affected and unaffected individuals, respectively.
The asterisk shows samples used for whole-exome sequencing. The mutations cosegregate with the phenotype. B: Schema of the CRYAA gene (top) and the CRYAA protein (bottom) is presented. The untranslated regions and coding region are shown as open and
filled rectangles, respectively. The location of the c.61C>T mutation is indicated with an arrow. CRYAA contains an N-terminal
region, an α-crystallin domain (ACD, dark gray box), and a C-terminal region. C: Electropherograms of the mutation in the affected patient (top) and the unaffected control (bottom) are shown. A single
nucleotide change in exon 1 results in an amino acid alteration. D: The missense mutation occurred at an evolutionarily conserved amino acid. Homologous sequences were aligned using CLUSTALW.
E: Schema of the CRYGC gene (top) and the CRYGC protein (bottom) is presented. The untranslated regions and the coding region are shown as open
and filled rectangles, respectively. The location of the c.124delT mutation is indicated with an arrow. CRYGC contains two
domains each composed of two Greek-key motifs (dark gray boxes). F: Electropherograms of the CRYGC mutation in the affected patient (top) and in the unaffected control (bottom) are shown.
A single nucleotide deletion in exon 2 would cause a frameshift. mut, mutant allele; wt, wild-type allele.
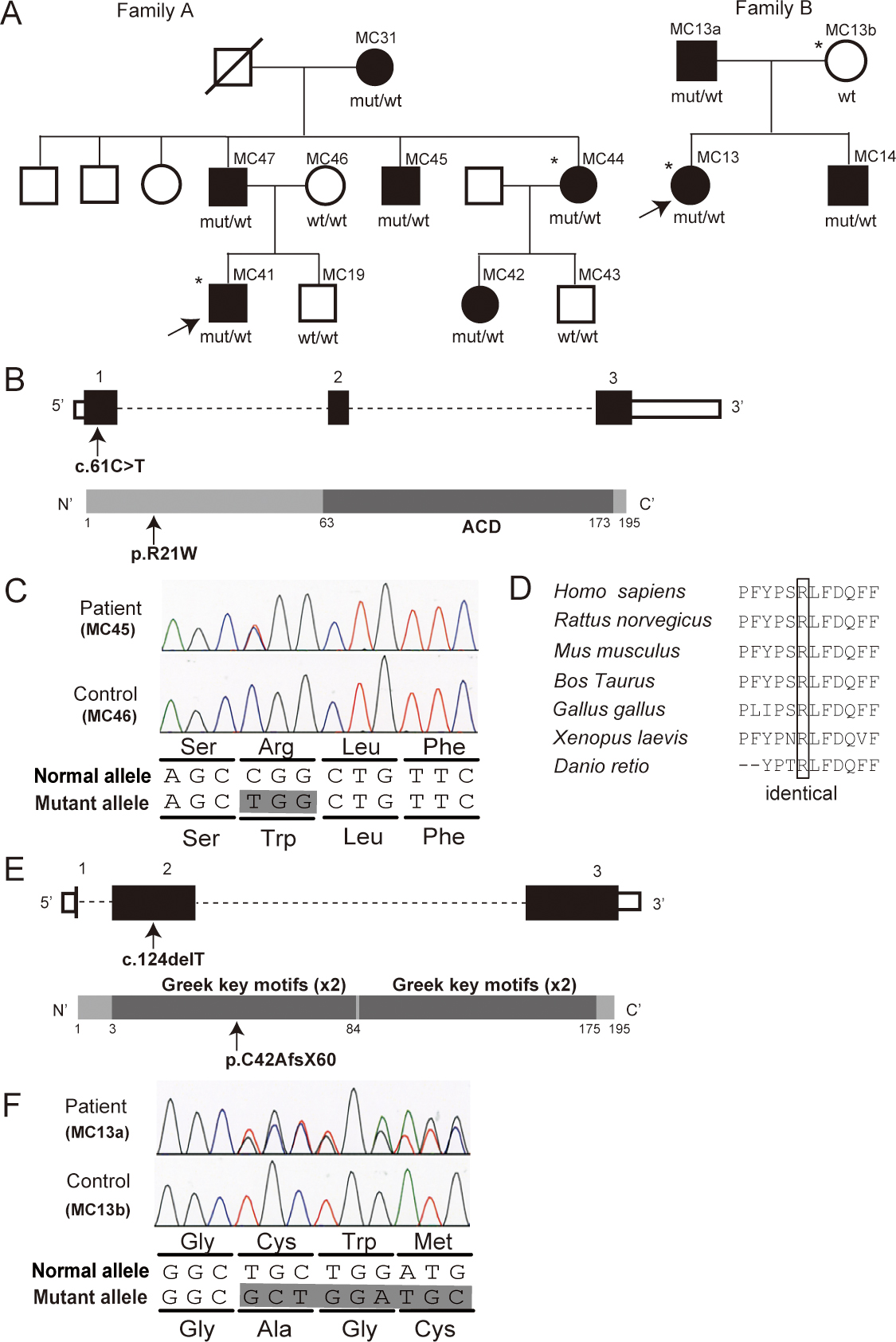
 Figure 1 of
Kondo, Mol Vis 2013; 19:384-389.
Figure 1 of
Kondo, Mol Vis 2013; 19:384-389.  Figure 1 of
Kondo, Mol Vis 2013; 19:384-389.
Figure 1 of
Kondo, Mol Vis 2013; 19:384-389. 