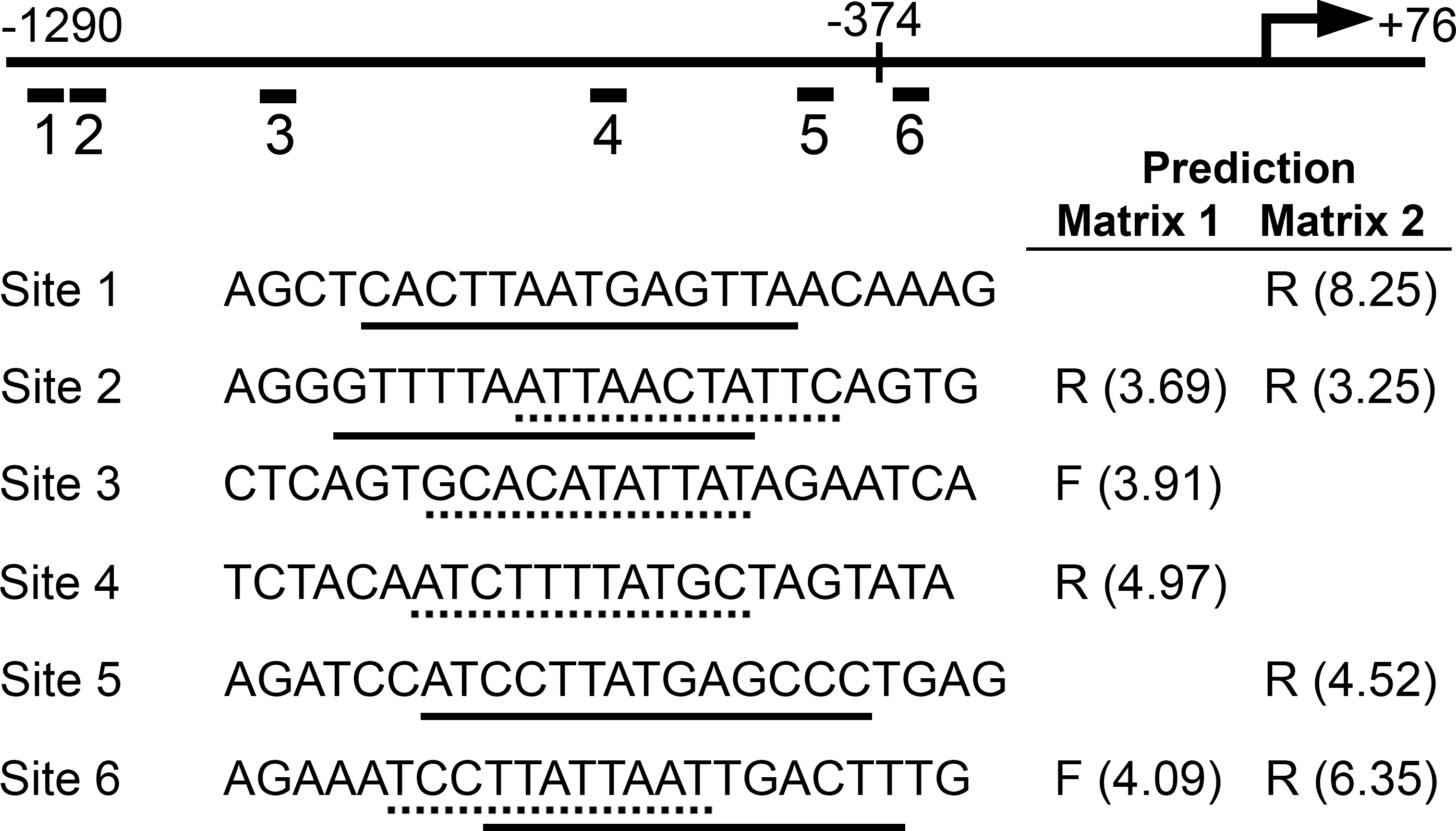
Figure 4. Bioinformatic prediction of POU4 protein binding sites. Nucleotide sequence of the
RIT2 upstream region from –1290 to +76 bp was scanned to search potential POU4 protein binding sites using the Patser program
with two position-specific scoring matrices, POU4F1/F2 matrix (Matrix 1) [
50] and POU4F2 matrix (Matrix 2) [
27]. The transcription start site (+1) is indicated with an angled arrow, and the position of –374 bp is marked with a vertical
line. Using 3.0 as a cut-off score, four sites were predicted with each matrix, and two sites (Sites 2 and 6) were predicted
with both matrices. The positions of the predicted sites (Sites 1–6) are indicated by short lines with numbers under the
RIT2 –1290 to +76 bp genomic segment. The sequences of oligonucleotides containing the predicted sites in the middle for EMSA
are shown. Core binding motifs predicted by Matrices 1 and 2 are marked with dotted and solid underlines, respectively (see
also Appendix 4). Strands of the predicted sites are also shown as F (forward) or R (reverse) with scores in parentheses.
 Figure 4 of
Zhang, Mol Vis 2013; 19:1371-1386.
Figure 4 of
Zhang, Mol Vis 2013; 19:1371-1386.  Figure 4 of
Zhang, Mol Vis 2013; 19:1371-1386.
Figure 4 of
Zhang, Mol Vis 2013; 19:1371-1386.